Structure, function, and substrates of Clp AAA+ protease systems in cyanobacteria, plastids, and apicoplasts: A comparative analysis
- PMID: 33497624
- PMCID: PMC7966870
- DOI: 10.1016/j.jbc.2021.100338
Structure, function, and substrates of Clp AAA+ protease systems in cyanobacteria, plastids, and apicoplasts: A comparative analysis
Erratum in
-
Correction: Structure, function, and substrates of Clp AAA+ protease systems in cyanobacteria, plastids, and apicoplasts: A comparative analysis.J Biol Chem. 2022 Mar;298(3):101705. doi: 10.1016/j.jbc.2022.101705. Epub 2022 Feb 13. J Biol Chem. 2022. PMID: 35168039 Free PMC article. No abstract available.
Abstract
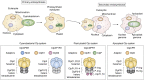
ATPases Associated with diverse cellular Activities (AAA+) are a superfamily of proteins that typically assemble into hexameric rings. These proteins contain AAA+ domains with two canonical motifs (Walker A and B) that bind and hydrolyze ATP, allowing them to perform a wide variety of different functions. For example, AAA+ proteins play a prominent role in cellular proteostasis by controlling biogenesis, folding, trafficking, and degradation of proteins present within the cell. Several central proteolytic systems (e.g., Clp, Deg, FtsH, Lon, 26S proteasome) use AAA+ domains or AAA+ proteins to unfold protein substrates (using energy from ATP hydrolysis) to make them accessible for degradation. This allows AAA+ protease systems to degrade aggregates and large proteins, as well as smaller proteins, and feed them as linearized molecules into a protease chamber. This review provides an up-to-date and a comparative overview of the essential Clp AAA+ protease systems in Cyanobacteria (e.g., Synechocystis spp), plastids of photosynthetic eukaryotes (e.g., Arabidopsis, Chlamydomonas), and apicoplasts in the nonphotosynthetic apicomplexan pathogen Plasmodium falciparum. Recent progress and breakthroughs in identifying Clp protease structures, substrates, substrate adaptors (e.g., NblA/B, ClpS, ClpF), and degrons are highlighted. We comment on the physiological importance of Clp activity, including plastid biogenesis, proteostasis, the chloroplast Protein Unfolding Response, and metabolism, across these diverse lineages. Outstanding questions as well as research opportunities and priorities to better understand the essential role of Clp systems in cellular proteostasis are discussed.
Keywords: AAA+ proteins; Clp protease; N-degrons; Proteases; apicoplasts; chloroplasts; plastids; protease adaptors; proteostasis.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Dohmen R.J., Wu P., Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. - PubMed
-
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
-
- Bouchnak I., van Wijk K.J. N-degron pathways in plastids. Trends Plant Sci. 2019;24:917–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

