Mechanisms Applied by Protein Inhibitors to Inhibit Cysteine Proteases
- PMID: 33498210
- PMCID: PMC7863939
- DOI: 10.3390/ijms22030997
Mechanisms Applied by Protein Inhibitors to Inhibit Cysteine Proteases
Abstract
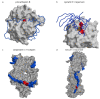
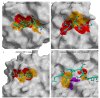
Protein inhibitors of proteases are an important tool of nature to regulate and control proteolysis in living organisms under physiological and pathological conditions. In this review, we analyzed the mechanisms of inhibition of cysteine proteases on the basis of structural information and compiled kinetic data. The gathered structural data indicate that the protein fold is not a major obstacle for the evolution of a protease inhibitor. It appears that nature can convert almost any starting fold into an inhibitor of a protease. In addition, there appears to be no general rule governing the inhibitory mechanism. The structural data make it clear that the "lock and key" mechanism is a historical concept with limited validity. However, the analysis suggests that the shape of the active site cleft of proteases imposes some restraints. When the S1 binding site is shaped as a pocket buried in the structure of protease, inhibitors can apply substrate-like binding mechanisms. In contrast, when the S1 binding site is in part exposed to solvent, the substrate-like inhibition cannot be employed. It appears that all proteases, with the exception of papain-like proteases, belong to the first group of proteases. Finally, we show a number of examples and provide hints on how to engineer protein inhibitors.
Keywords: compiled kinetic data; cysteine proteases inhibitors; mechanisms of inhibition; structural-based inhibition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Cryptostatin, a chagasin-family cysteine protease inhibitor of Cryptosporidium parvum.Parasitology. 2012 Jul;139(8):1029-37. doi: 10.1017/S0031182012000297. Epub 2012 Mar 23. Parasitology. 2012. PMID: 22444160
-
Molecular cloning of a multidomain cysteine protease and protease inhibitor precursor gene from the tobacco hornworm (Manduca sexta) and functional expression of the cathepsin F-like cysteine protease domain.Insect Biochem Mol Biol. 2010 Dec;40(12):835-46. doi: 10.1016/j.ibmb.2010.08.003. Epub 2010 Aug 18. Insect Biochem Mol Biol. 2010. PMID: 20727410
-
Cysteine protease inhibitor (AcStefin) is required for complete cyst formation of Acanthamoeba.Eukaryot Cell. 2013 Apr;12(4):567-74. doi: 10.1128/EC.00308-12. Epub 2013 Feb 8. Eukaryot Cell. 2013. PMID: 23397569 Free PMC article.
-
Microbial inhibitors of cysteine proteases.Med Microbiol Immunol. 2016 Aug;205(4):275-96. doi: 10.1007/s00430-016-0454-1. Epub 2016 Apr 5. Med Microbiol Immunol. 2016. PMID: 27048482 Review.
-
Papain-like lysosomal cysteine proteases and their inhibitors: drug discovery targets?Biochem Soc Symp. 2003;(70):15-30. doi: 10.1042/bss0700015. Biochem Soc Symp. 2003. PMID: 14587279 Review.
Cited by
-
The Role of Cysteine Protease Cathepsins B, H, C, and X/Z in Neurodegenerative Diseases and Cancer.Int J Mol Sci. 2023 Oct 26;24(21):15613. doi: 10.3390/ijms242115613. Int J Mol Sci. 2023. PMID: 37958596 Free PMC article. Review.
-
Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent.Biometals. 2024 Aug;37(4):983-1022. doi: 10.1007/s10534-024-00590-5. Epub 2024 Apr 5. Biometals. 2024. PMID: 38578560 Free PMC article.
-
Recovery of Proteases and Protease Inhibitors from Ganoderma spp. Cultivated in Amazonian Lignocellulose Wastes.Curr Protein Pept Sci. 2025;26(1):76-88. doi: 10.2174/0113892037297181240605112831. Curr Protein Pept Sci. 2025. PMID: 38919002
-
Emerging trends in the cystatin C sensing technologies: towards better chronic kidney disease management.RSC Adv. 2025 Feb 14;15(7):4926-4944. doi: 10.1039/d4ra07197b. eCollection 2025 Feb 13. RSC Adv. 2025. PMID: 39957820 Free PMC article. Review.
-
Trichocystatin-2 from Trichomonas vaginalis: role of N-terminal cysteines in aggregation, protease inhibition, and trichomonal cysteine protease-dependent cytotoxicity on HeLa cells.Front Parasitol. 2025 Mar 18;4:1512012. doi: 10.3389/fpara.2025.1512012. eCollection 2025. Front Parasitol. 2025. PMID: 40171250 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

