Human Transcriptomic Response to the VSV-Vectored Ebola Vaccine
- PMID: 33498214
- PMCID: PMC7908976
- DOI: 10.3390/vaccines9020067
Human Transcriptomic Response to the VSV-Vectored Ebola Vaccine
Abstract
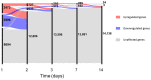
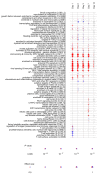
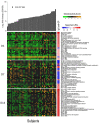
Ebolavirus Disease (EVD) is a severe haemorrhagic fever that occurs in epidemic outbreaks, with a high fatality rate and no specific therapies available. rVSVΔG-ZEBOV-GP (Ervebo®), a live-attenuated recombinant vesicular stomatitis virus vector expressing the glycoprotein G of Zaire Ebolavirus, is the first vaccine approved for prevention of EVD. Both innate and adaptive responses are deemed to be involved in vaccine-induced protection, yet the mechanisms are not fully elucidated. A global transcriptomic approach was used to profile the blood host-response in 51 healthy volunteers enrolled in a phase 1/2 clinical trial. Signatures of the host responses were investigated assessing the enrichment in differentially expressed genes (DEGs) of specific "blood transcription modules" (BTM). Comparison of gene-expression levels showed that vaccination produces a peak of 5469 DEGs at day one, representing 38.6% of the expressed genes. Out of 346 BTMs, 144 were significantly affected by vaccination. Innate immunity pathways were induced from day 1 to day 14. At days 2 and 3, neutrophil modules were downregulated and complement-related modules upregulated. T-cell and cell-cycle associated modules were upregulated at days 7 and 14, while at day 28, no modules remained activated. At day 14, a direct correlation was observed between ZEBOV glycoprotein-specific antibody titres and activation of seven BTMs, including two related to B-cell activation and B cell receptor signalling. Transcriptomic analysis identified an rVSVΔG-ZEBOV-GP-induced signature and demonstrated a direct correlation of blood transcriptomic changes with ZEBOV glycoprotein-specific antibody titres.
Keywords: Ebolavirus Disease; VSV-ZEBOV; live viral vector; recombinant VSV; transcriptomics; vaccine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Ebola Virus Disease. [(accessed on 7 December 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease.
-
- 2014–2016 Ebola Outbreak in West Africa|History|Ebola (Ebola Virus Disease)|CDC. [(accessed on 7 December 2020)]; Available online: https://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html.
-
- Ebola Health Update—North Kivu/Ituri, DRC, 2018–2020. [(accessed on 7 December 2020)]; Available online: https://www.who.int/emergencies/diseases/ebola/drc-2019.
-
- Vaccine against Ebola: Commission Grants Market Authorisation. [(accessed on 7 December 2020)]; Available online: https://ec.europa.eu/commission/presscorner/detail/en/ip_19_6246.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

