A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos
- PMID: 33498275
- PMCID: PMC7909245
- DOI: 10.3390/plants10020190
A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos
Abstract
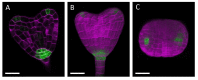
Tissue clearing methods combined with confocal microscopy have been widely used for studying developmental biology. In plants, ClearSee is a reliable clearing method that is applicable to a wide range of tissues and is suitable for gene expression analysis using fluorescent reporters, but its application to the Arabidopsis thaliana embryo, a model system to study morphogenesis and pattern formation, has not been described in the original literature. Here, we describe a ClearSee-based clearing protocol which is suitable for obtaining 3D images of Arabidopsis thaliana embryos. The method consists of embryo dissection, fixation, washing, clearing, and cell wall staining and enables high-quality 3D imaging of embryo morphology and expression of fluorescent reporters with the cellular resolution. Our protocol provides a reliable method that is applicable to the analysis of morphogenesis and gene expression patterns in Arabidopsis thaliana embryos.
Keywords: 3D imaging; Arabidopsis thaliana; GFP; cell wall staining; clearing; confocal microscopy; embryo; fluorescent reporter.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Scheres B., Wolkenfelt H., Willemsen V., Terlouw M., Lawson E., Dean C., Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–2487.
-
- Barton M.K., Poethig R.S. Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

