Towards Rational Biosurfactant Design-Predicting Solubilization in Rhamnolipid Solutions
- PMID: 33498574
- PMCID: PMC7864340
- DOI: 10.3390/molecules26030534
Towards Rational Biosurfactant Design-Predicting Solubilization in Rhamnolipid Solutions
Abstract
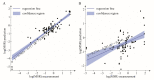
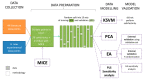
The efficiency of micellar solubilization is dictated inter alia by the properties of the solubilizate, the type of surfactant, and environmental conditions of the process. We, therefore, hypothesized that using the descriptors of the aforementioned features we can predict the solubilization efficiency, expressed as molar solubilization ratio (MSR). In other words, we aimed at creating a model to find the optimal surfactant and environmental conditions in order to solubilize the substance of interest (oil, drug, etc.). We focused specifically on the solubilization in biosurfactant solutions. We collected data from literature covering the last 38 years and supplemented them with our experimental data for different biosurfactant preparations. Evolutionary algorithm (EA) and kernel support vector machines (KSVM) were used to create predictive relationships. The descriptors of biosurfactant (logPBS, measure of purity), solubilizate (logPsol, molecular volume), and descriptors of conditions of the measurement (T and pH) were used for modelling. We have shown that the MSR can be successfully predicted using EAs, with a mean R2 val of 0.773 ± 0.052. The parameters influencing the solubilization efficiency were ranked upon their significance. This represents the first attempt in literature to predict the MSR with the MSR calculator delivered as a result of our research.
Keywords: MSR; QSAR; biosurfactant; efficiency; micellar solubilization; prediction; rhamnolipid.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Mulligan C.N., Yong R., Gibbs B. Surfactant-enhanced remediation of contaminated soil: A review. Eng. Geol. 2001;60:371–380. doi: 10.1016/S0013-7952(00)00117-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

