HIV-1 Gag-Pol Sequences from Ugandan Early Infections Reveal Sequence Variants Associated with Elevated Replication Capacity
- PMID: 33498793
- PMCID: PMC7912664
- DOI: 10.3390/v13020171
HIV-1 Gag-Pol Sequences from Ugandan Early Infections Reveal Sequence Variants Associated with Elevated Replication Capacity
Abstract
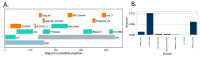
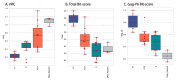
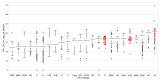
The ability to efficiently establish a new infection is a critical property for human immunodeficiency virus type 1 (HIV-1). Although the envelope protein of the virus plays an essential role in receptor binding and internalization of the infecting virus, the structural proteins, the polymerase and the assembly of new virions may also play a role in establishing and spreading viral infection in a new host. We examined Ugandan viruses from newly infected patients and focused on the contribution of the Gag-Pol genes to replication capacity. A panel of Gag-Pol sequences generated using single genome amplification from incident HIV-1 infections were cloned into a common HIV-1 NL4.3 pol/env backbone and the influence of Gag-Pol changes on replication capacity was monitored. Using a novel protein domain approach, we then documented diversity in the functional protein domains across the Gag-Pol region and identified differences in the Gag-p6 domain that were frequently associated with higher in vitro replication.
Keywords: Gag-Pol; HIV-1; Uganda; protein domains; recombinant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Fiebig E.W., Wright D.J., Rawal B.D., Garrett P.E., Schumacher R.T., Peddada L., Heldebrant C., Smith R., Conrad A., Kleinman S.H., et al. Dynamics of HIV Viremia and Antibody Seroconversion in Plasma Donors: Implications for Diagnosis and Staging of Primary HIV Infection. Aids. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. - DOI - PubMed
-
- Hansmann A., Schim van der Loeff M.F., Kaye S., Awasana A.A., Sarge-Njie R., O’Donovan D., Ariyoshi K., Alabi A., Milligan P., Whittle H.C. Baseline Plasma Viral Load and CD4 Cell Percentage Predict Survival in HIV-1- and HIV-2-Infected Women in a Community-Based Cohort in The Gambia. J. Acquir. Immune Defic. Syndr. 2005;38:335–341. - PubMed
-
- Price M.A., Rida W., Kilembe W., Karita E., Inambao M., Ruzagira E., Kamali A., Sanders E.J., Anzala O., Hunter E., et al. Control of the HIV-1 Load Varies by Viral Subtype in a Large Cohort of African Adults With Incident HIV-1 Infection. J. Infect. Dis. 2019;220:432–441. doi: 10.1093/infdis/jiz127. - DOI - PMC - PubMed
-
- Prentice H.A., Price M.A., Porter T.R., Cormier E., Mugavero M.J., Kamali A., Karita E., Lakhi S., Sanders E.J., Anzala O., et al. Dynamics of Viremia in Primary HIV-1 Infection in Africans: Insights from Analyses of Host and Viral Correlates. Virology. 2014;449:254–262. doi: 10.1016/j.virol.2013.11.024. - DOI - PMC - PubMed
-
- Huang W., Eshleman S.H., Toma J., Fransen S., Stawiski E., Paxinos E.E., Whitcomb J.M., Young A.M., Donnell D., Mmiro F., et al. Coreceptor Tropism in Human Immunodeficiency Virus Type 1 Subtype D: High Prevalence of CXCR4 Tropism and Heterogeneous Composition of Viral Populations. J. Virol. 2007;81:7885–7893. doi: 10.1128/JVI.00218-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

