Development of a Cell-Based Luciferase Complementation Assay for Identification of SARS-CoV-2 3CLpro Inhibitors
- PMID: 33498923
- PMCID: PMC7911889
- DOI: 10.3390/v13020173
Development of a Cell-Based Luciferase Complementation Assay for Identification of SARS-CoV-2 3CLpro Inhibitors
Abstract
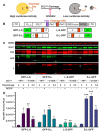
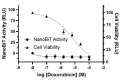
The 3C-like protease (3CLpro) of SARS-CoV-2 is considered an excellent target for COVID-19 antiviral drug development because it is essential for viral replication and has a cleavage specificity distinct from human proteases. However, drug development for 3CLpro has been hindered by a lack of cell-based reporter assays that can be performed in a BSL-2 setting. Current efforts to identify 3CLpro inhibitors largely rely upon in vitro screening, which fails to account for cell permeability and cytotoxicity of compounds, or assays involving replication-competent virus, which must be performed in a BSL-3 facility. To address these limitations, we have developed a novel cell-based luciferase complementation reporter assay to identify inhibitors of SARS-CoV-2 3CLpro in a BSL-2 setting. The assay is based on a lentiviral vector that co-expresses 3CLpro and two luciferase fragments linked together by a 3CLpro cleavage site. 3CLpro-mediated cleavage results in a loss of complementation and low luciferase activity, whereas inhibition of 3CLpro results in 10-fold higher levels of luciferase activity. The luciferase reporter assay can easily distinguish true 3CLpro inhibition from cytotoxicity, a powerful feature that should reduce false positives during screening. Using the assay, we screened 32 small molecules for activity against SARS-CoV-2 3CLpro, including HIV protease inhibitors, HCV protease inhibitors, and various other compounds that have been reported to inhibit SARS-CoV-2 3CLpro. Of these, only five exhibited significant inhibition of 3CLpro in cells: GC376, boceprevir, Z-FA-FMK, calpain inhibitor XII, and GRL-0496. This assay should greatly facilitate efforts to identify more potent inhibitors of SARS-CoV-2 3CLpro.
Keywords: 3CLpro; COVID-19; SARS-CoV-2; antiviral; inhibitor; luciferase; protease.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

