Autophagy-A Story of Bacteria Interfering with the Host Cell Degradation Machinery
- PMID: 33499114
- PMCID: PMC7911818
- DOI: 10.3390/pathogens10020110
Autophagy-A Story of Bacteria Interfering with the Host Cell Degradation Machinery
Abstract
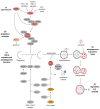
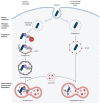
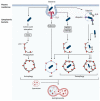
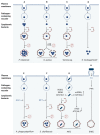
Autophagy is a highly conserved and fundamental cellular process to maintain cellular homeostasis through recycling of defective organelles or proteins. In a response to intracellular pathogens, autophagy further acts as an innate immune response mechanism to eliminate pathogens. This review will discuss recent findings on autophagy as a reaction to intracellular pathogens, such as Salmonella typhimurium, Listeria monocytogenes, Mycobacterium tuberculosis, Staphylococcus aureus, and pathogenic Escherichia coli. Interestingly, while some of these bacteria have developed methods to use autophagy for their own benefit within the cell, others have developed fascinating mechanisms to evade recognition, to subvert the autophagic pathway, or to escape from autophagy.
Keywords: autophagy; innate immune response; pathogens; pattern recognition receptors; xenophagy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Salmonella Suppresses the TRIF-Dependent Type I Interferon Response in Macrophages.mBio. 2016 Feb 16;7(1):e02051-15. doi: 10.1128/mBio.02051-15. mBio. 2016. PMID: 26884434 Free PMC article.
-
The role of autophagy in bacterial infections.Biosci Trends. 2015 Jun;9(3):149-59. doi: 10.5582/bst.2015.01035. Biosci Trends. 2015. PMID: 26166368 Review.
-
Survival of intracellular pathogens in response to mTORC1- or TRPML1-TFEB-induced xenophagy.Autophagy Rep. 2023 Mar 19;2(1):2191918. doi: 10.1080/27694127.2023.2191918. eCollection 2023. Autophagy Rep. 2023. PMID: 40395298 Free PMC article.
-
Bacterial Autophagy: Offense and Defense at the Host-Pathogen Interface.Cell Mol Gastroenterol Hepatol. 2017 May 6;4(2):237-243. doi: 10.1016/j.jcmgh.2017.05.002. eCollection 2017 Sep. Cell Mol Gastroenterol Hepatol. 2017. PMID: 28660242 Free PMC article. Review.
-
Autophagy targeting of Listeria monocytogenes and the bacterial countermeasure.Autophagy. 2011 Mar;7(3):310-4. doi: 10.4161/auto.7.3.14581. Autophagy. 2011. PMID: 21193840 Review.
Cited by
-
Mycoplasma bovis inhibits autophagy in bovine mammary epithelial cells via a PTEN/PI3K-Akt-mTOR-dependent pathway.Front Microbiol. 2022 Jul 26;13:935547. doi: 10.3389/fmicb.2022.935547. eCollection 2022. Front Microbiol. 2022. PMID: 35958147 Free PMC article.
-
When the Phagosome Gets Leaky: Pore-Forming Toxin-Induced Non-Canonical Autophagy (PINCA).Front Cell Infect Microbiol. 2022 Mar 17;12:834321. doi: 10.3389/fcimb.2022.834321. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35372127 Free PMC article.
-
DRAM1 Promotes Lysosomal Delivery of Mycobacterium marinum in Macrophages.Cells. 2023 Mar 7;12(6):828. doi: 10.3390/cells12060828. Cells. 2023. PMID: 36980169 Free PMC article.
-
Programmed cell death and melatonin: A comprehensive review.Funct Integr Genomics. 2024 Sep 24;24(5):169. doi: 10.1007/s10142-024-01454-4. Funct Integr Genomics. 2024. PMID: 39313718 Review.
-
Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy.Int J Mol Sci. 2021 Nov 18;22(22):12466. doi: 10.3390/ijms222212466. Int J Mol Sci. 2021. PMID: 34830349 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

