Sex-Stratified Gene Regulatory Networks Reveal Female Key Driver Genes of Atherosclerosis Involved in Smooth Muscle Cell Phenotype Switching
- PMID: 33499648
- PMCID: PMC7930467
- DOI: 10.1161/CIRCULATIONAHA.120.051231
Sex-Stratified Gene Regulatory Networks Reveal Female Key Driver Genes of Atherosclerosis Involved in Smooth Muscle Cell Phenotype Switching
Abstract
Background: Although sex differences in coronary artery disease are widely accepted with women developing more stable atherosclerosis than men, the underlying pathobiology of such differences remains largely unknown. In coronary artery disease, recent integrative systems biological studies have inferred gene regulatory networks (GRNs). Within these GRNs, key driver genes have shown great promise but have thus far been unidentified in women.
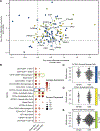
Methods: We generated sex-specific GRNs of the atherosclerotic arterial wall in 160 women and age-matched men in the STARNET study (Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task). We integrated the female GRNs with single-cell RNA-sequencing data of the human atherosclerotic plaque and single-cell RNA sequencing of advanced atherosclerotic lesions from wild type and Klf4 knockout atherosclerotic smooth muscle cell (SMC) lineage-tracing mice.
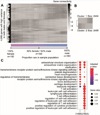
Results: By comparing sex-specific GRNs, we observed clear sex differences in network activity within the atherosclerotic tissues. Genes more active in women were associated with mesenchymal cells and endothelial cells, whereas genes more active in men were associated with the immune system. We determined that key drivers of GRNs active in female coronary artery disease were predominantly found in (SMCs by single-cell sequencing of the human atherosclerotic plaques, and higher expressed in female plaque SMCs, as well. To study the functions of these female SMC key drivers in atherosclerosis, we examined single-cell RNA sequencing of advanced atherosclerotic lesions from wild type and Klf4 knockout atherosclerotic SMC lineage-tracing mice. The female key drivers were found to be expressed by phenotypically modulated SMCs and affected by Klf4, suggesting that sex differences in atherosclerosis involve phenotypic switching of plaque SMCs.
Conclusions: Our systems approach provides novel insights into molecular mechanisms that underlie sex differences in atherosclerosis. To discover sex-specific therapeutic targets for atherosclerosis, an increased emphasis on sex-stratified approaches in the analysis of multi-omics data sets is warranted.
Keywords: atherosclerosis; gene regulatory networks; myocytes, smooth muscle; sex; systems biology; transcriptome.
Conflict of interest statement
Conflict of interest:
There are no conflicts of interest to disclose.
Figures




References
-
- Vrijenhoek JEP, Haitjema S, De Borst GJ, De Vries JPPM, Vaartjes I, Moll FL, Pasterkamp G, Den Ruijter HM. The impact of female sex on long-term survival of patients with severe atherosclerosis undergoing endarterectomy. Atherosclerosis. 2014;237:521–527. - PubMed
-
- Vrijenhoek JEP, Den Ruijter HM, De Borst GJ, De Kleijn DP V, De Vries JPPM, Bots ML, Van De Weg SM, Vink A, Moll FL, Pasterkamp G. Sex is associated with the presence of atherosclerotic plaque hemorrhage and modifies the relation between plaque hemorrhage and cardiovascular outcome. Stroke. 2013;44:3318–3323. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

