Mammalian BCAS3 and C16orf70 associate with the phagophore assembly site in response to selective and non-selective autophagy
- PMID: 33499712
- PMCID: PMC8386740
- DOI: 10.1080/15548627.2021.1874133
Mammalian BCAS3 and C16orf70 associate with the phagophore assembly site in response to selective and non-selective autophagy
Abstract
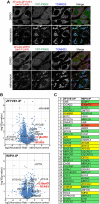
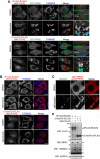
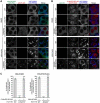
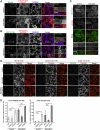
Macroautophagy/autophagy is an intracellular degradation process that delivers cytosolic materials and/or damaged organelles to lysosomes. De novo synthesis of the autophagosome membrane occurs within a phosphatidylinositol-3-phosphate-rich region of the endoplasmic reticulum, and subsequent expansion is critical for cargo encapsulation. This process is complex, especially in mammals, with many regulatory factors. In this study, by utilizing PRKN (parkin RBR E3 ubiquitin protein ligase)-mediated mitochondria autophagy (mitophagy)-inducing conditions in conjunction with chemical crosslinking and mass spectrometry, we identified human BCAS3 (BCAS3 microtubule associated cell migration factor) and C16orf70 (chromosome 16 open reading frame 70) as novel proteins that associate with the autophagosome formation site during both non-selective and selective autophagy. We demonstrate that BCAS3 and C16orf70 form a complex and that their association with the phagophore assembly site requires both proteins. In silico structural modeling, mutational analyses in cells and in vitro phosphoinositide-binding assays indicate that the WD40 repeat domain in human BCAS3 directly binds phosphatidylinositol-3-phosphate. Furthermore, overexpression of the BCAS3-C16orf70 complex affects the recruitment of several core autophagy proteins to the phagophore assembly site. This study demonstrates regulatory roles for human BCAS3 and C16orf70 in autophagic activity.Abbreviations: AO: antimycin A and oligomycin; Ash: assembly helper; ATG: autophagy-related; BCAS3: BCAS3 microtubule associated cell migration factor; C16orf70: chromosome 16 open reading frame 70; DAPI: 4',6-diamidino-2-phenylindole; DKO: double knockout; DMSO: dimethyl sulfoxide; ER: endoplasmic reticulum; fluoppi: fluorescent-based technology detecting protein-protein interactions; FIS1: fission, mitochondrial 1; FKBP: FKBP prolyl isomerase family member 1C; FRB: FKBP-rapamycin binding; hAG: humanized azami-green; IP: immunoprecipitation; IRES: internal ribosome entry site; KO: knockout; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MFN2: mitofusin 2; MS: mass spectrometry; MT-CO2: mitochondrially encoded cytochrome c oxidase II; mtDNA: mitochondrial DNA; OPTN: optineurin; PFA: paraformaldehyde; PE: phosphatidylethanolamine; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; PtdIns(3,5)P2: phosphatidylinositol-3,5-bisphosphate; PINK1: PTEN induced kinase 1; PRKN/Parkin: parkin RBR E3 ubiquitin protein ligase; PROPPIN: β-propellers that bind polyphosphoinositides; RB1CC1/FIP200: RB1 inducible coiled-coil 1; TOMM20: translocase of outer mitochondrial membrane 20; ULK1: unc-51 like autophagy activating kinase 1; WDR45B/WIPI3: WD repeat domain 45B; WDR45/WIPI4: WD repeat domain 45; WIPI: WD repeat domain, phosphoinositide interacting; WT: wild type; ZFYVE1/DFCP1: zinc finger FYVE-type containing 1.
Keywords: Mitophagy; parkin; phagophore; pink1; starvation; wd40.
Conflict of interest statement
No competing interests declared
Figures






References
-
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
