Antibiotic resistance profiles in cultivable microbiota isolated from some romanian natural fishery lakes included in Natura 2000 network
- PMID: 33499841
- PMCID: PMC7836572
- DOI: 10.1186/s12917-021-02770-8
Antibiotic resistance profiles in cultivable microbiota isolated from some romanian natural fishery lakes included in Natura 2000 network
Abstract
Background: The present study aims the characterization of antibiotic resistance phenotypes and encoding genes in bacterial strains isolated from some Romanian aquatic fishery lowland salted lakes.
Material/methods: This study was conducted on 44 bacterial strains, mainly belonging to species used as microbiological indicators of fecal pollution isolated from four natural fishery lakes. All strains were tested for their antibiotic susceptibility by disk diffusion method. Simplex and multiplex PCR were performed to identify the β-lactams antibiotic resistance genes (blaNMD, blaOXA-48, blaVIM, blaIMP, blaCTX-M, blaTEM), sulfonamides (Sul1, Sul2), tetracyclines (TetA, TetB, TetC, TetD, TetM), aminoglycosides (aac3Ia), vancomycin (VanA, VanB, VanC), macrolides (ermA, ermB, ermC) as well as the plasmid-mediated quinolone resistance (PMQR) markers (QnrA, QnrB, QnrS), and class 1 integrons (Int1, drfA1-aadA1).
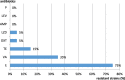
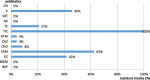
Results: The Enterococcus spp. isolates exhibited phenotypic resistance to vancomycin (35 %) and macrolides (erythromycin) (75 %); from the vancomycin - resistant strains, 5 % harboured VanA (E. faecalis), while the erythromycin resistant isolates were positive for the ermA gene (E. faecalis - 10 %, E. faecium - 5 %). The Gram- negative rods (GNR) exhibited a high level of resistance to β-lactams: cefuroxime (63 %), cefazolin (42 %), ceftriaxone (8 %), ceftazidime and aztreonam (4 % each). The genetic determinants for beta-lactam resistance were represented by blaCTX-M-like (33 %), blaNDM-like and blaIMP-like (8.33 %) genes. The resistance to non-β-lactam antibiotics was ascertained to the following genes: quinolones (QnrS - 4.16 %); sulfonamides (Sul1-75 %, Sul2-4.16 %); aminoglycosides (aac3Ia - 4.16 %); tetracyclines (tetA - 25 %, tetC - 15 %). The integrase gene was found in more than 50 % of the studied strains (58.33 %).
Conclusions: The cultivable aquatic microbiota from fishery lakes is dominated by enterococci and Enterobacterales strains. The GNR strains exhibited high levels of β-lactam resistance mediated by extended spectrum beta-lactamases and metallo-β-lactamases. The Enterococcus sp. isolates were highly resistant to macrolides and vancomycin. The high level and diversity of resistance markers, correlated with a high frequency of integrons is suggesting that this environment could act as an important reservoir of antibiotic resistance genes with a great probability to be horizontally transmitted to other associated species from the aquatic sediments microbiota, raising the potential zoonotic risk for fish consumers.
Keywords: Antimicrobial resistance; Enterobacterales; Enterococcus sp.; Lowland fishery lakes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Zhang XX, Zhang T, Fang HHP. Appl Microbiol B. 2009. 82:397 https://www.ecdc.europa.eu/en/antimicrobial-consumption/database/country.... Accesed at 10.06.2020;.
-
- Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall AB, Wellington EM. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011;5(8):1253–61. doi: 10.1038/ismej.2011.15. - DOI - PMC - PubMed
-
- González-Zorn B, Escudero JA. Ecology of antimicrobial resistance: humans, animals, food and environment. Int Microbiol. 2012;15(3):101–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

