Role of human Pegivirus infections in whole Plasmodium falciparum sporozoite vaccination and controlled human malaria infection in African volunteers
- PMID: 33499880
- PMCID: PMC7837505
- DOI: 10.1186/s12985-021-01500-8
Role of human Pegivirus infections in whole Plasmodium falciparum sporozoite vaccination and controlled human malaria infection in African volunteers
Abstract
Background: Diverse vaccination outcomes and protection levels among different populations pose a serious challenge to the development of an effective malaria vaccine. Co-infections are among many factors associated with immune dysfunction and sub-optimal vaccination outcomes. Chronic, asymptomatic viral infections can contribute to the modulation of vaccine efficacy through various mechanisms. Human Pegivirus-1 (HPgV-1) persists in immune cells thereby potentially modulating immune responses. We investigated whether Pegivirus infection influences vaccine-induced responses and protection in African volunteers undergoing whole P. falciparum sporozoites-based malaria vaccination and controlled human malaria infections (CHMI).
Methods: HPgV-1 prevalence was quantified by RT-qPCR in plasma samples of 96 individuals before, post vaccination with PfSPZ Vaccine and after CHMI in cohorts from Tanzania and Equatorial Guinea. The impact of HPgV-1 infection was evaluated on (1) systemic cytokine and chemokine levels measured by Luminex, (2) PfCSP-specific antibody titers quantified by ELISA, (3) asexual blood-stage parasitemia pre-patent periods and parasite multiplication rates, (4) HPgV-1 RNA levels upon asexual blood-stage parasitemia induced by CHMI.
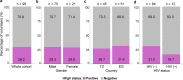
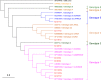
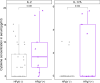
Results: The prevalence of HPgV-1 was 29.2% (28/96) and sequence analysis of the 5' UTR and E2 regions revealed the predominance of genotypes 1, 2 and 5. HPgV-1 infection was associated with elevated systemic levels of IL-2 and IL-17A. Comparable vaccine-induced anti-PfCSP antibody titers, asexual blood-stage multiplication rates and pre-patent periods were observed in HPgV-1 positive and negative individuals. However, a tendency for higher protection levels was detected in the HPgV-1 positive group (62.5%) compared to the negative one (51.6%) following CHMI. HPgV-1 viremia levels were not significantly altered after CHMI.
Conclusions: HPgV-1 infection did not alter PfSPZ Vaccine elicited levels of PfCSP-specific antibody responses and parasite multiplication rates. Ongoing HPgV-1 infection appears to improve to some degree protection against CHMI in PfSPZ-vaccinated individuals. This is likely through modulation of immune system activation and systemic cytokines as higher levels of IL-2 and IL17A were observed in HPgV-1 infected individuals. CHMI is safe and well tolerated in HPgV-1 infected individuals. Identification of cell types and mechanisms of both silent and productive infection in individuals will help to unravel the biology of this widely present but largely under-researched virus.
Keywords: Antibody response; Controlled human malaria infection; Human pegivirus; Immune activation; Malaria; PfSPZ vaccine.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Controlled Human Malaria Infection of Healthy Adults With Lifelong Malaria Exposure to Assess Safety, Immunogenicity, and Efficacy of the Asexual Blood Stage Malaria Vaccine Candidate GMZ2.Clin Infect Dis. 2019 Sep 27;69(8):1377-1384. doi: 10.1093/cid/ciy1087. Clin Infect Dis. 2019. PMID: 30561539 Free PMC article. Clinical Trial.
-
Attenuated PfSPZ Vaccine induces strain-transcending T cells and durable protection against heterologous controlled human malaria infection.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2711-2716. doi: 10.1073/pnas.1615324114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223498 Free PMC article. Clinical Trial.
-
PfSPZ Vaccine induces focused humoral immune response in HIV positive and negative Tanzanian adults.EBioMedicine. 2024 Oct;108:105364. doi: 10.1016/j.ebiom.2024.105364. Epub 2024 Sep 30. EBioMedicine. 2024. PMID: 39353279 Free PMC article.
-
Protective efficacy and safety of radiation-attenuated and chemo-attenuated Plasmodium Falciparum sporozoite vaccines against controlled and natural malaria infection: a systematic review and meta-analysis of randomized controlled trials.Infection. 2024 Jun;52(3):707-722. doi: 10.1007/s15010-024-02174-4. Epub 2024 Feb 6. Infection. 2024. PMID: 38319556
-
Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines.Vaccine. 2015 Dec 22;33(52):7452-61. doi: 10.1016/j.vaccine.2015.09.096. Epub 2015 Nov 27. Vaccine. 2015. PMID: 26469720 Free PMC article. Review.
Cited by
-
Dramatic transcriptomic differences in Macaca mulatta and Macaca fascicularis with Plasmodium knowlesi infections.Sci Rep. 2021 Sep 30;11(1):19519. doi: 10.1038/s41598-021-98024-6. Sci Rep. 2021. PMID: 34593836 Free PMC article.
-
Human pegivirus alters brain and blood immune and transcriptomic profiles of patients with Parkinson's disease.JCI Insight. 2025 Jul 8;10(13):e189988. doi: 10.1172/jci.insight.189988. eCollection 2025 Jul 8. JCI Insight. 2025. PMID: 40626361 Free PMC article.
-
Review of human pegivirus: Prevalence, transmission, pathogenesis, and clinical implication.Virulence. 2022 Dec;13(1):324-341. doi: 10.1080/21505594.2022.2029328. Virulence. 2022. PMID: 35132924 Free PMC article. Review.
References
-
- World malaria report 2019. https://www.who.int/publications-detail/world-malaria-report-2019. Accessed May 20, 2020
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

