Community-curated and standardised metadata of published ancient metagenomic samples with AncientMetagenomeDir
- PMID: 33500403
- PMCID: PMC7838265
- DOI: 10.1038/s41597-021-00816-y
Community-curated and standardised metadata of published ancient metagenomic samples with AncientMetagenomeDir
Abstract
Ancient DNA and RNA are valuable data sources for a wide range of disciplines. Within the field of ancient metagenomics, the number of published genetic datasets has risen dramatically in recent years, and tracking this data for reuse is particularly important for large-scale ecological and evolutionary studies of individual taxa and communities of both microbes and eukaryotes. AncientMetagenomeDir (archived at https://doi.org/10.5281/zenodo.3980833 ) is a collection of annotated metagenomic sample lists derived from published studies that provide basic, standardised metadata and accession numbers to allow rapid data retrieval from online repositories. These tables are community-curated and span multiple sub-disciplines to ensure adequate breadth and consensus in metadata definitions, as well as longevity of the database. Internal guidelines and automated checks facilitate compatibility with established sequence-read archives and term-ontologies, and ensure consistency and interoperability for future meta-analyses. This collection will also assist in standardising metadata reporting for future ancient metagenomic studies.
Conflict of interest statement
The authors declare no competing interests.
Figures
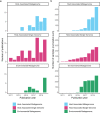
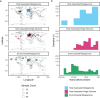

References
Publication types
MeSH terms
Grants and funding
- 208934/Z/17/Z/Wellcome Trust (Wellcome)/International
- EXC 2051-Project-ID 390713860/Deutsche Forschungsgemeinschaft (German Research Foundation)/International
- T32 GM007197/GM/NIGMS NIH HHS/United States
- DGE1255832/National Science Foundation (NSF)/International
- ERC-2014-ADG 670518/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

