A new SYBR Green real-time PCR to detect SARS-CoV-2
- PMID: 33500453
- PMCID: PMC7838253
- DOI: 10.1038/s41598-021-81245-0
A new SYBR Green real-time PCR to detect SARS-CoV-2
Abstract
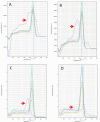
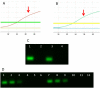
Phylogenetic analysis has demonstrated that the etiologic agent of the 2020 pandemic outbreak is a betacoronavirus named SARS-CoV-2. For public health interventions, a diagnostic test with high sensitivity and specificity is required. The gold standard protocol for diagnosis by the Word Health Organization (WHO) is RT-PCR. To detect low viral loads and perform large-scale screening, a low-cost diagnostic test is necessary. Here, we developed a cost-effective test capable of detecting SARS-CoV-2. We validated an auxiliary protocol for molecular diagnosis with the SYBR Green RT-PCR methodology to successfully screen negative cases of SARS-CoV-2. Our results revealed a set of primers with high specificity and no homology with other viruses from the Coronovideae family or human respiratory tract pathogenic viruses, presenting with complementarity only for rhinoviruses/enteroviruses and Legionella spp. Optimization of the annealing temperature and polymerization time led to a high specificity in the PCR products. We have developed a more affordable and swift methodology for negative SARS-CoV-2 screening. This methodology can be applied on a large scale to soften panic and economic burden through guidance for isolation strategies.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

