Parental nucleosome segregation and the inheritance of cellular identity
- PMID: 33500558
- PMCID: PMC8609916
- DOI: 10.1038/s41576-020-00312-w
Parental nucleosome segregation and the inheritance of cellular identity
Abstract
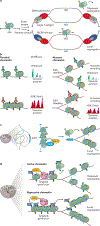
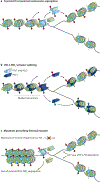
Gene expression programmes conferring cellular identity are achieved through the organization of chromatin structures that either facilitate or impede transcription. Among the key determinants of chromatin organization are the histone modifications that correlate with a given transcriptional status and chromatin state. Until recently, the details for the segregation of nucleosomes on DNA replication and their implications in re-establishing heritable chromatin domains remained unclear. Here, we review recent findings detailing the local segregation of parental nucleosomes and highlight important advances as to how histone methyltransferases associated with the establishment of repressive chromatin domains facilitate epigenetic inheritance.
Conflict of interest statement
Competing interests
D.R. is a co-founder of Constellation Pharmaceuticals and Fulcrum Therapeutics. The other authors declare no competing interests.
Figures


References
-
- Waddington CH The epigenotype. 1942. Int. J. Epidemiol 41, 10–13 (2012). - PubMed
-
- Allis CD, Caparros M-L, Jenuwein T, Reinberg D & Lachner M Epigenetics (Cold Spring Harbor Laboratory Press, 2015).
-
- Kornberg RD Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 (1974). - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

