Systematic large-scale assessment of the genetic architecture of left ventricular noncompaction reveals diverse etiologies
- PMID: 33500567
- PMCID: PMC8105165
- DOI: 10.1038/s41436-020-01049-x
Systematic large-scale assessment of the genetic architecture of left ventricular noncompaction reveals diverse etiologies
Abstract
Purpose: To characterize the genetic architecture of left ventricular noncompaction (LVNC) and investigate the extent to which it may represent a distinct pathology or a secondary phenotype associated with other cardiac diseases.
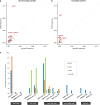
Methods: We performed rare variant association analysis with 840 LVNC cases and 125,748 gnomAD population controls, and compared results to similar analyses on dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM).
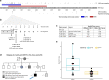
Results: We observed substantial genetic overlap indicating that LVNC often represents a phenotypic variation of DCM or HCM. In contrast, truncating variants in MYH7, ACTN2, and PRDM16 were uniquely associated with LVNC and may reflect a distinct LVNC etiology. In particular, MYH7 truncating variants (MYH7tv), generally considered nonpathogenic for cardiomyopathies, were 20-fold enriched in LVNC cases over controls. MYH7tv heterozygotes identified in the UK Biobank and healthy volunteer cohorts also displayed significantly greater noncompaction compared with matched controls. RYR2 exon deletions and HCN4 transmembrane variants were also enriched in LVNC, supporting prior reports of association with arrhythmogenic LVNC phenotypes.
Conclusion: LVNC is characterized by substantial genetic overlap with DCM/HCM but is also associated with distinct noncompaction and arrhythmia etiologies. These results will enable enhanced application of LVNC genetic testing and help to distinguish pathological from physiological noncompaction.
Conflict of interest statement
S.A.C. is a cofounder and director of Enleofen Bio PTE Ltd, a company that develops anti-IL-11 therapeutics. Enleofen Bio had no involvement in this study. J.S.W. and I.O. receive grant support and honoraria from Myokardia. Myokardia had no involvement in this study. The other authors declare no competing interests.
Figures
References
-
- Hershberger, R. E., Morales, A. & Cowan, J. Is left ventricular noncompaction a trait, phenotype, or disease? The evidence points to phenotype. Circ. Cardiovasc. Genet.10, e001968 (2017). - PubMed
-
- Maron BJ, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functio. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

