Androgen receptor gain in circulating free DNA and splicing variant 7 in exosomes predict clinical outcome in CRPC patients treated with abiraterone and enzalutamide
- PMID: 33500577
- PMCID: PMC8134038
- DOI: 10.1038/s41391-020-00309-w
Androgen receptor gain in circulating free DNA and splicing variant 7 in exosomes predict clinical outcome in CRPC patients treated with abiraterone and enzalutamide
Abstract
Background: Androgen receptor (AR) signaling inhibitors represent the standard treatment in metastatic castration resistance prostate cancer (mCRPC) patients. However, some patients display a primary resistance, and several studies investigated the role of the AR as a predictive biomarker of response to treatment. This study is aimed to evaluate the role of AR in liquid biopsy to predict clinical outcome to AR signaling inhibitors in mCRPC patients.
Methods: Six milliliters of plasma samples were collected before first-line treatment with abiraterone or enzalutamide. Circulating free DNA (cfDNA) and exosome-RNA were isolated for analysis of AR gain and AR splice variant 7 (AR-V7), respectively, by digital droplet PCR.
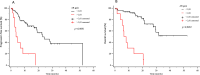
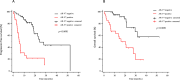
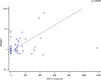
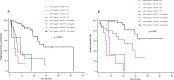
Results: Eighty-four mCRPC patients received abiraterone (n = 40) or enzalutamide (n = 44) as first-line therapy. Twelve patients (14.3%) presented AR gain and 30 (35.7%) AR-V7+ at baseline. Median progression-free survival (PFS) and overall survival (OS) were significantly longer in AR-V7- vs AR-V7+ patients (24.3 vs 5.4 months, p < 0.0001; not reached vs 16.2 months, p = 0.0001, respectively). Patients carrying the AR gain had a median PFS of 4.8 vs 24.3 months for AR normal patients (p < 0.0001). Median OS was significantly longer in AR normal vs patients with AR gain (not reached vs 8.17 months, p < 0.0001). A significant correlation between AR-V7 and AR gain was observed (r = 0.28; p = 0.01). The AR gain/AR-V7 combined analysis confirmed a strong predictive effect for biomarkers combination vs patients without any AR aberration (PFS 3.8 vs 28 month, respectively; OS 6.1 vs not reached, respectively; p < 0.0001).
Conclusions: The present study demonstrates that cfDNA and exosome-RNA are both a reliable source of AR variants and their combined detection in liquid biopsy predicts resistance to AR signaling inhibitors.
Conflict of interest statement
MDR received speaker honoraria from Astellas, Astra Zeneca, Celgene, Novartis, Pfizer, Bio-Rad Janssen-Cilag, Sanofi-Aventis; consulting fee from Ipsen and Janssen-Cilag; Speaker’s bureau: Celgene, Janssen, Sanofi; travel support from Janssen, Bio-Rad. VC has received speaker honoraria or travel support from Astellas, Janssen-Cilag, and Sanofi-Aventis, and has received consulting fee from Bayer. RD received honoraria for scientific advisory board and consulting relationship from Ipsen, Novartis, Pfizer, Sanofi Genzyme, AstraZeneca, Janssen, Gilead, Lilly, Gilead, EUSA Pharma; travel support from Ipsen, Sanofi Genzyme. UDG received honoraria as consulting or advisory role for Pfizer, Janssen, Astellas Pharma, Sanofi, Bristol-Myers Squibb, Bayer, Ipsen, Merck; received institutional research funding from Sanofi, AstraZeneca, Roche; received travel support from Bristol-Myers Squibb, Ipsen, Janssen, Pfizer. All remaining authors have declared no conflicts of interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

