High LARGE1 Expression May Predict Benefit from Adjuvant Chemotherapy in Resected Non-Small-Cell Lung Cancer
- PMID: 33500650
- PMCID: PMC7822230
- DOI: 10.2147/PGPM.S271516
High LARGE1 Expression May Predict Benefit from Adjuvant Chemotherapy in Resected Non-Small-Cell Lung Cancer
Abstract
Background: LARGE1 plays a pivotal role in glycosylation of alpha-Dystroglycan (α-DG) and is aberrantly downregulated in cell lines originating from epithelium-derived cancers including lung cancer. However, the expression of LARGE1 and its clinical significance in NSCLC are not clear.
Materials and methods: The data were collected from the TCGA database to investigate LARGE1 expression in stage I-III NSCLC and explore its associations with clinicopathological parameters and overall survival of patients. The prognostic role of LARGE1 was examined in subgroups according to clinical features and treatments. The results were validated in external cohorts from the NCBI GEO database. Gene Set Enrichment Analysis (GSEA) was performed to investigate the potential molecular mechanisms during LARGE1 alteration in NSCLC.
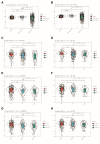
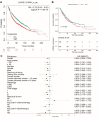
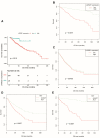
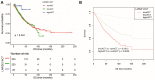
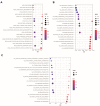
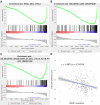
Results: LARGE1 was aberrantly downregulated in NSCLC compared with adjacent tissues and normal lung tissues and in tumors with advanced stage compared with early stage. There was only a trend of association between high LARGE1 with OS in multivariate analysis. Surprisingly, high LARGE1 was significantly associated with improved OS in a subgroup of the patients with adjuvant chemotherapy (ACT) and a significant interaction between LARGE1 expression and ACT was found. Improved OS after ACT was also found in patients with high LARGE1 compared to those with low LARGE1. When combining LARGE1 expression and ACT, compared with patients with non-ACT, HR of low LARGE1/ACT was 0.592 (95% CI=0.432-0.813, P=0.0012), and HR of high LARGE1/ACT was 0.124 (95% CI=0.031-0.505, P=0.0036). The results were verified in two external cohorts from the GEO database. GSEA indicated that LARGE1 might downregulate cell cycle pathway to improve ACT sensitivity and subsequently the prognosis in NSCLC.
Conclusion: High LARGE1 can be used to identify the patients with resected stage I-III NSCLC most likely to benefit from adjuvant chemotherapy.
Keywords: LARGE1; NSCLC; adjuvant chemotherapy; benefit; mRNA; overall survival.
© 2021 Liu et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):425. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

