Advances and Perspectives in Applying Deep Learning for Drug Design and Discovery
- PMID: 33501123
- PMCID: PMC7805776
- DOI: 10.3389/frobt.2019.00108
Advances and Perspectives in Applying Deep Learning for Drug Design and Discovery
Abstract
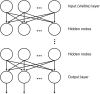
Discovering (or planning) a new drug candidate involves many parameters, which makes this process slow, costly, and leading to failures at the end in some cases. In the last decades, we have witnessed a revolution in the computational area (hardware, software, large-scale computing, etc.), as well as an explosion in data generation (big data), which raises the need for more sophisticated algorithms to analyze this myriad of data. In this scenario, we can highlight the potentialities of artificial intelligence (AI) or computational intelligence (CI) as a powerful tool to analyze medicinal chemistry data. According to IEEE, computational intelligence involves the theory, the design, the application, and the development of biologically and linguistically motivated computational paradigms. In addition, CI encompasses three main methodologies: neural networks (NN), fuzzy systems, and evolutionary computation. In particular, artificial neural networks have been successfully applied in medicinal chemistry studies. A branch of the NN area that has attracted a lot of attention refers to deep learning (DL) due to its generalization power and ability to extract features from data. Therefore, in this mini-review we will briefly outline the present scope, advances, and challenges related to the use of DL in drug design and discovery, describing successful studies involving quantitative structure-activity relationships (QSAR) and virtual screening (VS) of databases containing thousands of compounds.
Keywords: artificial intelligence; deep learning; drug design; drug discovery; medicinal chemistry.
Copyright © 2019 Lipinski, Maltarollo, Oliveira, da Silva and Honorio.
Figures

References
-
- Capuzzi S. J., Politi R., Isayev O., Farag S., Tropsha A. (2016). QSAR modeling of Tox21 challenge stress response and nuclear receptor signaling toxicity assays. Front. Environ. Sci. 4:3 10.3389/fenvs.2016.00003 - DOI
-
- Caruana R. (1997). Multitask learning. Mach. Learn. 28, 41–75. 10.1023/A:1007379606734 - DOI
-
- Chen J., Jin Q., Chao J. (2012). Design of deep belief networks for short-term prediction of drought index using data in the Huaihe River basin. Math. Probl. Eng. 2012:16 10.1155/2012/235929 - DOI
Publication types
LinkOut - more resources
Full Text Sources

