This is a preprint.
SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma
- PMID: 33501446
- PMCID: PMC7836116
- DOI: 10.1101/2021.01.18.427166
SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma
Update in
-
SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma.Nat Med. 2021 Apr;27(4):622-625. doi: 10.1038/s41591-021-01285-x. Epub 2021 Mar 2. Nat Med. 2021. PMID: 33654292
Abstract
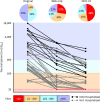
SARS-CoV-2 501Y.V2 (B.1.351), a novel lineage of coronavirus causing COVID-19, contains substitutions in two immunodominant domains of the spike protein. Here, we show that pseudovirus expressing 501Y.V2 spike protein completely escapes three classes of therapeutically relevant antibodies. This pseudovirus also exhibits substantial to complete escape from neutralization, but not binding, by convalescent plasma. These data highlight the prospect of reinfection with antigenically distinct variants and foreshadows reduced efficacy of spike-based vaccines.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures

References
-
- Marovich M., Mascola J.R. & Cohen M.S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA 324, 131–132 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
