Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish
- PMID: 33502030
- PMCID: PMC8653899
- DOI: 10.1111/mec.15817
Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish
Abstract
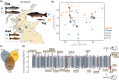
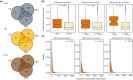
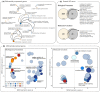
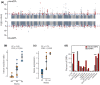
Understanding the contribution of different molecular processes to evolution and development is crucial for identifying the mechanisms of adaptation. Here, we used RNA-sequencing data to test the importance of alternative splicing and differential gene expression in a case of parallel adaptive evolution, the replicated postglacial divergence of the salmonid fish Arctic charr (Salvelinus alpinus) into sympatric benthic and pelagic ecotypes across multiple independent lakes. We found that genes differentially spliced between ecotypes were mostly not differentially expressed (<6% overlap) and were involved in different biological processes. Differentially spliced genes were primarily enriched for muscle development and functioning, while differentially expressed genes were involved in metabolism, immunity and growth. Furthermore, alternative splicing and gene expression were mostly controlled by independent cis-regulatory quantitative trait loci (<3.4% overlap). Cis-regulatory regions were associated with the parallel divergence in splicing (16.5% of intron clusters) and expression (6.7%-10.1% of differentially expressed genes), indicating shared regulatory variation across ecotype pairs. Contrary to theoretical expectation, we found that differentially spliced genes tended to be highly central in regulatory networks ("hub genes") and were annotated to significantly more gene ontology terms compared to nondifferentially spliced genes, consistent with a higher level of pleiotropy. Together, our results suggest that the concerted regulation of alternative splicing and differential gene expression through different regulatory regions leads to the divergence of complementary processes important for local adaptation. This provides novel insights into the importance of contrasting but putatively complementary molecular processes in rapid parallel adaptive evolution.
Keywords: adaptive divergence; alternative splicing; convergent evolution; ecological speciation; gene expression; parallel evolution; salmonid.
© 2021 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Figures

Comment in
-
Alternative splicing: An overlooked mechanism contributing to local adaptation?Mol Ecol. 2021 Oct;30(20):4951-4954. doi: 10.1111/mec.16177. Epub 2021 Oct 6. Mol Ecol. 2021. PMID: 34533856
Similar articles
-
Alternative splicing: An overlooked mechanism contributing to local adaptation?Mol Ecol. 2021 Oct;30(20):4951-4954. doi: 10.1111/mec.16177. Epub 2021 Oct 6. Mol Ecol. 2021. PMID: 34533856
-
Parallelism in eco-morphology and gene expression despite variable evolutionary and genomic backgrounds in a Holarctic fish.PLoS Genet. 2020 Apr 17;16(4):e1008658. doi: 10.1371/journal.pgen.1008658. eCollection 2020 Apr. PLoS Genet. 2020. PMID: 32302300 Free PMC article.
-
The interplay between epigenomic and transcriptomic variation during ecotype divergence in stickleback.BMC Biol. 2025 Mar 5;23(1):70. doi: 10.1186/s12915-025-02176-0. BMC Biol. 2025. PMID: 40038570 Free PMC article.
-
Environmental correlates of adaptive diversification in postglacial freshwater fishes.J Fish Biol. 2024 Mar;104(3):517-535. doi: 10.1111/jfb.15621. Epub 2023 Dec 10. J Fish Biol. 2024. PMID: 37984834 Review.
-
Genetic Causes and Consequences of Sympatric Morph Divergence in Salmonidae: A Search for Mechanisms.Annu Rev Anim Biosci. 2022 Feb 15;10:81-106. doi: 10.1146/annurev-animal-051021-080709. Epub 2021 Nov 10. Annu Rev Anim Biosci. 2022. PMID: 34758272 Review.
Cited by
-
The roles of different gene expression regulators in acoustic variation in the intermediate horseshoe bat revealed by long-read and short-read RNA sequencing data.Curr Zool. 2023 Sep 30;70(5):575-588. doi: 10.1093/cz/zoad045. eCollection 2024 Oct. Curr Zool. 2023. PMID: 39463690 Free PMC article.
-
Alternative splicing perspective to prey preference of environmentally friendly biological agent Cryptolaemus montrouzieri.BMC Genomics. 2024 Oct 15;25(1):967. doi: 10.1186/s12864-024-10870-6. BMC Genomics. 2024. PMID: 39407100 Free PMC article.
-
Alternative Splicing Variation: Accessing and Exploiting in Crop Improvement Programs.Int J Mol Sci. 2023 Oct 15;24(20):15205. doi: 10.3390/ijms242015205. Int J Mol Sci. 2023. PMID: 37894886 Free PMC article. Review.
-
Application of Omics Tools in Designing and Monitoring Marine Protected Areas For a Sustainable Blue Economy.Front Genet. 2022 Jun 22;13:886494. doi: 10.3389/fgene.2022.886494. eCollection 2022. Front Genet. 2022. PMID: 35812740 Free PMC article.
-
Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain.Microorganisms. 2025 Jan 14;13(1):165. doi: 10.3390/microorganisms13010165. Microorganisms. 2025. PMID: 39858933 Free PMC article.
References
-
- Adams, C. E. , & Huntingford, F. A. (2002). Inherited differences in head allometry in polymorphic Arctic charr from Loch Rannoch, Scotland. Journal of Fish Biology, 60(3), 515–520.
-
- Adams, C. E. , & Huntingford, F. A. (2002). The functional significance of inherited differences in feeding morphology in a sympatric polymorphic population of Arctic charr. Evolutionary Ecology, 16(1), 15–25.
-
- Adams, C. E. , & Huntingford, F. A. (2004). Incipient speciation driven by phenotypic plasticity? Evidence from sympatric populations of Arctic charr. Biological Journal of the Linnean Society. Linnean Society of London, 81(4), 611–618.
-
- Alekseyev, S. S. , Samusenok, V. P. , Matveev, A. N. , & Pichugin, M. Y. (2002). Diversification, sympatric speciation, and trophic polymorphism of arctic Charr, Salvelinus Alpinus Complex. Transbaikalia. Environmental Biology of Fishes, 64(1), 97–114.
-
- Altringham, J. D. , & Ellerby, D. J. (1999). Fish swimming: Patterns in muscle function. The Journal of Experimental Biology, 202(Pt 23), 3397–3403. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

