Colonization with Staphylococcus aureus and Klebsiella pneumoniae causes infections in a Vietnamese intensive care unit
- PMID: 33502303
- PMCID: PMC8208697
- DOI: 10.1099/mgen.0.000514
Colonization with Staphylococcus aureus and Klebsiella pneumoniae causes infections in a Vietnamese intensive care unit
Abstract
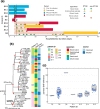
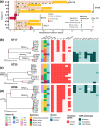
Pre-existing colonization with Staphylococcus aureus or Klebsiella pneumoniae has been found to increase the risk of infection in intensive care patients. We previously conducted a longitudinal study to characterize colonization of these two organisms in patients admitted to intensive care in a hospital in southern Vietnam. Here, using genomic and phylogenetic analyses, we aimed to assess the contribution these colonizing organisms made to infections. We found that in the majority of patients infected with S. aureus or K. pneumoniae, the sequence type of the disease-causing (infecting) isolate was identical to that of corresponding colonizing organisms in the respective patient. Further in-depth analysis revealed that in patients infected by S. aureus ST188 and by K. pneumoniae ST17, ST23, ST25 and ST86, the infecting isolate was closely related to and exhibited limited genetic variation relative to pre-infection colonizing isolates. Multidrug-resistant S. aureus ST188 was identified as the predominant agent of colonization and infection. Colonization and infection by K. pneumoniae were characterized by organisms with limited antimicrobial resistance profiles but extensive repertoires of virulence genes. Our findings augment the understanding of the link between bacterial colonization and infection in a low-resource setting, and could facilitate the development of novel evidence-based approaches to prevent and treat infections in high-risk patients in intensive care.
Keywords: Staphylococcus aureus; colonization; hospital-acquired infections; hypervirulent Klebsiella pneumoniae; intra-patient diversity.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures

References
-
- WHO Report on the burden of endemic health care-associated infection worldwide clean care is safer care. World Heal Organ. 2011:1–40.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

