A non-radioactive, improved PAR-CLIP and small RNA cDNA library preparation protocol
- PMID: 33503264
- PMCID: PMC8096255
- DOI: 10.1093/nar/gkab011
A non-radioactive, improved PAR-CLIP and small RNA cDNA library preparation protocol
Abstract
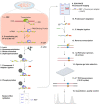
Crosslinking and immunoprecipitation (CLIP) methods are powerful techniques to interrogate direct protein-RNA interactions and dissect posttranscriptional gene regulatory networks. One widely used CLIP variant is photoactivatable ribonucleoside enhanced CLIP (PAR-CLIP) that involves in vivo labeling of nascent RNAs with the photoreactive nucleosides 4-thiouridine (4SU) or 6-thioguanosine (6SG), which can efficiently crosslink to interacting proteins using UVA and UVB light. Crosslinking of 4SU or 6SG to interacting amino acids changes their base-pairing properties and results in characteristic mutations in cDNA libraries prepared for high-throughput sequencing, which can be computationally exploited to remove abundant background from non-crosslinked sequences and help pinpoint RNA binding protein binding sites at nucleotide resolution on a transcriptome-wide scale. Here we present a streamlined protocol for fluorescence-based PAR-CLIP (fPAR-CLIP) that eliminates the need to use radioactivity. It is based on direct ligation of a fluorescently labeled adapter to the 3'end of crosslinked RNA on immobilized ribonucleoproteins, followed by isolation of the adapter-ligated RNA and efficient conversion into cDNA without the previously needed size fractionation on denaturing polyacrylamide gels. These improvements cut the experimentation by half to 2 days and increases sensitivity by 10-100-fold.
Published by Oxford University Press on behalf of Nucleic Acids Research 2021.
Figures


References
-
- Ray D., Kazan H., Chan E.T., Pena Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R.. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009; 27:667–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

