Aspergillus fumigatus tryptophan metabolic route differently affects host immunity
- PMID: 33503414
- PMCID: PMC7844877
- DOI: 10.1016/j.celrep.2020.108673
Aspergillus fumigatus tryptophan metabolic route differently affects host immunity
Abstract
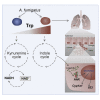
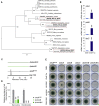
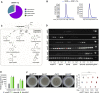
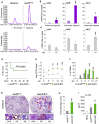
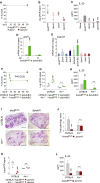
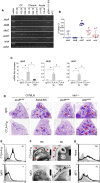
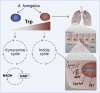
Indoleamine 2,3-dioxygenases (IDOs) degrade l-tryptophan to kynurenines and drive the de novo synthesis of nicotinamide adenine dinucleotide. Unsurprisingly, various invertebrates, vertebrates, and even fungi produce IDO. In mammals, IDO1 also serves as a homeostatic regulator, modulating immune response to infection via local tryptophan deprivation, active catabolite production, and non-enzymatic cell signaling. Whether fungal Idos have pleiotropic functions that impact on host-fungal physiology is unclear. Here, we show that Aspergillus fumigatus possesses three ido genes that are expressed under conditions of hypoxia or tryptophan abundance. Loss of these genes results in increased fungal pathogenicity and inflammation in a mouse model of aspergillosis, driven by an alternative tryptophan degradation pathway to indole derivatives and the host aryl hydrocarbon receptor. Fungal tryptophan metabolic pathways thus cooperate with the host xenobiotic response to shape host-microbe interactions in local tissue microenvironments.
Keywords: AhR; Aspergillus fumigatus; IDO; IL-33; NAD; aspergillosis; indoles; inflammation; tryptophan.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Agus A., Planchais J., Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

