Metabolic compensation activates pro-survival mTORC1 signaling upon 3-phosphoglycerate dehydrogenase inhibition in osteosarcoma
- PMID: 33503424
- PMCID: PMC8552368
- DOI: 10.1016/j.celrep.2020.108678
Metabolic compensation activates pro-survival mTORC1 signaling upon 3-phosphoglycerate dehydrogenase inhibition in osteosarcoma
Abstract
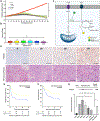
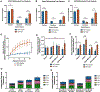
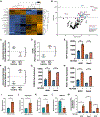
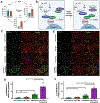
Osteosarcoma is the most common pediatric and adult primary malignant bone cancer. Curative regimens target the folate pathway, downstream of serine metabolism, with high-dose methotrexate. Here, the rate-limiting enzyme in the biosynthesis of serine from glucose, 3-phosphoglycerate dehydrogenase (PHGDH), is examined, and an inverse correlation between PHGDH expression and relapse-free and overall survival in osteosarcoma patients is found. PHGDH inhibition in osteosarcoma cell lines attenuated cellular proliferation without causing cell death, prompting a robust metabolic analysis to characterize pro-survival compensation. Using metabolomic and lipidomic profiling, cellular response to PHGDH inhibition is identified as accumulation of unsaturated lipids, branched chain amino acids, and methionine cycle intermediates, leading to activation of pro-survival mammalian target of rapamycin complex 1 (mTORC1) signaling. Increased mTORC1 activation sensitizes cells to mTORC1 pathway inhibition, resulting in significant, synergistic cell death in vitro and in vivo. Identifying a therapeutic combination for PHGDH-high cancers offers preclinical justification for a dual metabolism-based combination therapy for osteosarcoma.
Keywords: GATOR; PHGDH; SAMTOR; lipid metabolism; mTORC1; methotrexate; one-carbon metabolism; osteosarcoma; perhexiline; serine biosynthesis.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.C.P. declares current employment at Aveda. B.A.V.T. declares grants from Merck; grants and personal fees from Pfizer; grants from TRACON Pharmaceuticals; grants, personal fees, and other remunerations from GlaxoSmithKline; personal fees from Polaris Inc.; personal fees from Lilly; personal fees from Caris Life Sciences; personal fees from Novartis; personal fees from CytRX; personal fees from Plexxikon; personal fees from Epizyme; personal fees from Daiichi Sankyo; personal fees from Adaptimmune; personal fees from Immune Design; personal fees from Bayer; personal fees from Cytokinetics; and personal fees from Deciphera; he has a patent issued for the use of ME1 as a biomarker and ACXT3102. The other authors declare no competing interests.
Figures


References
-
- Beadle RM, Williams LK, Kuehl M, Bowater S, Abozguia K, Leyva F, Yousef Z, Wagenmakers AJM, Thies F, Horowitz J, and Frenneaux MP (2015). Improvement in cardiac energetics by perhexiline in heart failure due to dilated cardiomyopathy. JACC Heart Fail. 3, 202–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

