Nicotinic cholinergic system and COVID-19: In silico identification of interactions between α7 nicotinic acetylcholine receptor and the cryptic epitopes of SARS-Co-V and SARS-CoV-2 Spike glycoproteins
- PMID: 33503469
- PMCID: PMC7830272
- DOI: 10.1016/j.fct.2021.112009
Nicotinic cholinergic system and COVID-19: In silico identification of interactions between α7 nicotinic acetylcholine receptor and the cryptic epitopes of SARS-Co-V and SARS-CoV-2 Spike glycoproteins
Abstract
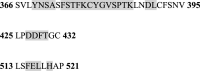
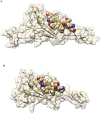
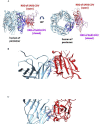
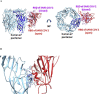
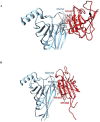
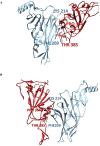
SARS-CoV-2 is the coronavirus that originated in Wuhan in December 2019 and has spread globally. Studies have shown that smokers are less likely to be diagnosed with or be hospitalized for COVID-19 but, once hospitalized, have higher odds for an adverse outcome. We have previously presented the potential interaction between SARS-CoV-2 Spike glycoprotein and nicotinic acetylcholine receptors (nAChRs), due to a "toxin-like" epitope on the Spike glycoprotein, with homology to a sequence of a snake venom toxin. This epitope coincides with the well-described cryptic epitope for the human anti-SARS-CoV antibody CR3022. In this study, we present the molecular complexes of both SARS-CoV and SARS-CoV-2 Spike glycoproteins, at their open or closed conformations, with the model of the human α7 nAChR. We found that all studied protein complexes' interface involves a large part of the "toxin-like" sequences of SARS-CoV and SARS-CoV-2 Spike glycoproteins and toxin binding site of human α7 nAChR. Our findings provide further support to the hypothesis about the protective role of nicotine and other cholinergic agonists. The potential therapeutic role of CR3022 and other similar monoclonal antibodies with increased affinity for SARS-CoV-2 Spike glycoprotein against the clinical effects originating from the dysregulated cholinergic pathway should be further explored.
Keywords: COVID-19; CR3022; Cryptic epitope; Molecular modeling; SARS-CoV-2; Spike glycoprotein; nAChRs.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Socrates J. Tzartos, Elias Eliopoulos, Konstantinos Poulas and Konstantinos Farsalinos are listed as inventors on pending patent application for cholinergic agonists and anti-SPIKE antibodies.
Figures



Similar articles
-
Nicotinic Cholinergic System and COVID-19: In Silico Identification of an Interaction between SARS-CoV-2 and Nicotinic Receptors with Potential Therapeutic Targeting Implications.Int J Mol Sci. 2020 Aug 13;21(16):5807. doi: 10.3390/ijms21165807. Int J Mol Sci. 2020. PMID: 32823591 Free PMC article.
-
Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.MAbs. 2021 Jan-Dec;13(1):1953683. doi: 10.1080/19420862.2021.1953683. MAbs. 2021. PMID: 34313527 Free PMC article.
-
SARS-CoV-2 spike ectodomain targets α7 nicotinic acetylcholine receptors.J Biol Chem. 2023 May;299(5):104707. doi: 10.1016/j.jbc.2023.104707. Epub 2023 Apr 13. J Biol Chem. 2023. PMID: 37061001 Free PMC article.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
Nicotine and the nicotinic cholinergic system in COVID-19.FEBS J. 2020 Sep;287(17):3656-3663. doi: 10.1111/febs.15521. Epub 2020 Aug 25. FEBS J. 2020. PMID: 32790936 Free PMC article. Review.
Cited by
-
Myasthenia Gravis Presenting after Administration of the mRNA-1273 Vaccine.Eur J Case Rep Intern Med. 2022 Jul 12;9(7):003439. doi: 10.12890/2022_003439. eCollection 2022. Eur J Case Rep Intern Med. 2022. PMID: 36051160 Free PMC article.
-
A Deadly Embrace: Hemagglutination Mediated by SARS-CoV-2 Spike Protein at Its 22 N-Glycosylation Sites, Red Blood Cell Surface Sialoglycoproteins, and Antibody.Int J Mol Sci. 2022 Feb 25;23(5):2558. doi: 10.3390/ijms23052558. Int J Mol Sci. 2022. PMID: 35269703 Free PMC article. Review.
-
Smoking Status and Factors associated with COVID-19 In-Hospital Mortality among US Veterans.Nicotine Tob Res. 2022 Mar 26;24(5):785-793. doi: 10.1093/ntr/ntab223. Nicotine Tob Res. 2022. PMID: 34693967 Free PMC article.
-
Maternal COVID-19 Vaccination and Its Potential Impact on Fetal and Neonatal Development.Vaccines (Basel). 2021 Nov 18;9(11):1351. doi: 10.3390/vaccines9111351. Vaccines (Basel). 2021. PMID: 34835282 Free PMC article. Review.
-
Could Small Neurotoxins-Peptides be Expressed during SARS-CoV-2 Infection?Curr Genomics. 2021 Dec 31;22(8):557-563. doi: 10.2174/1389202923666211221111527. Curr Genomics. 2021. PMID: 35382352 Free PMC article.
References
-
- Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P., Placid Trial Collaborators Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020 Oct 22:371. doi: 10.1136/bmj.m3939. m3939. - DOI - PMC - PubMed
-
- Alqahtani J.S., Oyelade T., Aldhahir A.M., Alghamdi S.M., Almehmadi M., Alqahtani A.S., Quaderi S., Mandal S., Hurst J.R. Prevalence, severity and mortality associated with COPD and smoking in patients with COVID-19: a rapid systematic review and meta-analysis. PloS One. 2020 May 11;15(5) doi: 10.1371/journal.pone.0233147. - DOI - PMC - PubMed
-
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., et al. Identification of neutralizing human monoclonal antibodies from Italian Covid-19 convalescent patients. bioRxiv. 2020 doi: 10.1101/2020.05.05.078154. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

