Cardiomyocytes Derived from Induced Pluripotent Stem Cells as a Disease Model for Propionic Acidemia
- PMID: 33503868
- PMCID: PMC7865492
- DOI: 10.3390/ijms22031161
Cardiomyocytes Derived from Induced Pluripotent Stem Cells as a Disease Model for Propionic Acidemia
Abstract
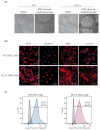
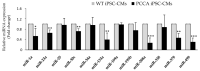
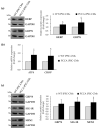
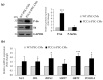
Propionic acidemia (PA), one of the most frequent life-threatening organic acidemias, is caused by mutations in either the PCCA or PCCB genes encoding both subunits of the mitochondrial propionyl-CoA carboxylase (PCC) enzyme. Cardiac alterations (hypertrophy, dilated cardiomyopathy, long QT) are one of the major causes of mortality in patients surviving the neonatal period. To overcome limitations of current cellular models of PA, we generated induced pluripotent stem cells (iPSCs) from a PA patient with defects in the PCCA gene, and successfully differentiated them into cardiomyocytes. PCCA iPSC-derived cardiomyocytes exhibited reduced oxygen consumption, an accumulation of residual bodies and lipid droplets, and increased ribosomal biogenesis. Furthermore, we found increased protein levels of HERP, GRP78, GRP75, SIG-1R and MFN2, suggesting endoplasmic reticulum stress and calcium perturbations in these cells. We also analyzed a series of heart-enriched miRNAs previously found deregulated in the heart tissue of a PA murine model and confirmed their altered expression. Our novel results show that PA iPSC-cardiomyocytes represent a promising model for investigating the pathological mechanisms underlying PA cardiomyopathies, also serving as an ex vivo platform for therapeutic evaluation.
Keywords: cardiac dysfunction; disease model; iPSC; iPSC-derived cardiomyocytes; propionic acidemia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

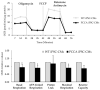
References
-
- Fenton W.A., Gravel R.A., Rosenberg L.E. Disorders of propionate and methylmalonate metabolism. In: Scriver C.R., Beaudet A.L., Sly W., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. 8th ed. McGraw-Hill; New York, NY, USA: 2001. pp. 2165–2190.
MeSH terms
Substances
Grants and funding
- PAF107/Propionic Acidemia Foundation
- SAF2016-76004-R/Spanish Ministry of Economy, Industry and Competitiveness - Agencia Estatal de Investigación and European Regional Development Fund
- PID2019-105344RB-I00/AEI/10.13039/501100011033/Spanish Ministry of Science and Innovation
- LCF/PR/PR16/11110018/Fundación Isabel Gemio, Fundación La Caixa
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

