Genomic Characterization of Clinical Extensively Drug-Resistant Acinetobacter pittii Isolates
- PMID: 33503968
- PMCID: PMC7912037
- DOI: 10.3390/microorganisms9020242
Genomic Characterization of Clinical Extensively Drug-Resistant Acinetobacter pittii Isolates
Abstract
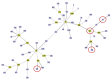
Carbapenem-resistant Acinetobacter pittii (CRAP) is a causative agent of nosocomial infections. This study aimed to characterize clinical isolates of CRAP from a tertiary hospital in Northeast Thailand. Six isolates were confirmed as extensively drug-resistant Acinetobacter pittii (XDRAP). The blaNDM-1 gene was detected in three isolates, whereas blaIMP-14 and blaIMP-1 were detected in the others. Multilocus sequence typing with the Pasteur scheme revealed ST220 in two isolates, ST744 in two isolates, and ST63 and ST396 for the remaining two isolates, respectively. Genomic characterization revealed that six XDRAP genes contained antimicrobial resistance genes: ST63 (A436) and ST396 (A1) contained 10 antimicrobial resistance genes, ST220 (A984 and A864) and ST744 (A56 and A273) contained 9 and 8 antimicrobial resistance genes, respectively. The single nucleotide polymorphism (SNP) phylogenetic tree revealed that the isolates A984 and A864 were closely related to A. pittii YB-45 (ST220) from China, while A436 was related to A. pittii WCHAP100020, also from China. A273 and A56 isolates (ST744) were clustered together; these isolates were closely related to strains 2014S07-126, AP43, and WCHAP005069, which were isolated from Taiwan and China. Strict implementation of infection control based upon the framework of epidemiological analyses is essential to prevent outbreaks and contain the spread of the pathogen. Continued surveillance and close monitoring with molecular epidemiological tools are needed.
Keywords: Acinetobacter pittii; Thailand; XDR; carbapenem-resistance; extensively drug-resistant; whole genome sequencing.
Conflict of interest statement
The authors have no conflicts of interest in this study.
Figures


References
-
- Nemec A., Krizova L., Maixnerova M., van der Reijden T.J.K., Deschaght P., Passet V., Vaneechoutte M., Brisse S., Dijkshoorn L. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov.(formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov.(formerly Acinetobacter genomic species 13TU) Res. Microbiol. 2011;162:393–404. doi: 10.1016/j.resmic.2011.02.006. - DOI - PubMed
-
- Nemec A., Krizova L., Maixnerova M., Sedo O., Brisse S., Higgins P.G. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2015;65:934–942. doi: 10.1099/ijs.0.000043. - DOI - PubMed
-
- Cosgaya C., Marí-Almirall M., van Assche A., Assche V., Fern Andez-Orth D., Mosqueda N., Telli M., Huys G., Higgins P.G., Seifert H., et al. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 2016;66:4105–4111. doi: 10.1099/ijsem.0.001318. - DOI - PubMed
-
- Iimura M., Hayashi W., Arai E., Natori T., Horiuchi K., Go M., Tanaka H., Soga E., Nagano Y., Nagano N. Detection of Acinetobacter pittii ST220 co-producing NDM-1 and OXA-820 carbapenemases from a hospital sink in a non-endemic country of NDM. J. Glob. Antimicrob. Resist. 2019 doi: 10.1016/j.jgar.2019.11.013. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

