Molecular Transmission Dynamics of Primary HIV Infections in Lazio Region, Years 2013-2020
- PMID: 33503987
- PMCID: PMC7911907
- DOI: 10.3390/v13020176
Molecular Transmission Dynamics of Primary HIV Infections in Lazio Region, Years 2013-2020
Abstract
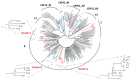
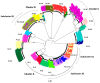
Molecular investigation of primary HIV infections (PHI) is crucial to describe current dynamics of HIV transmission. Aim of the study was to investigate HIV transmission clusters (TC) in PHI referred during the years 2013-2020 to the National Institute for Infectious Diseases in Rome (INMI), that is the Lazio regional AIDS reference centre, and factors possibly associated with inclusion in TC. These were identified by phylogenetic analysis, based on population sequencing of pol; a more in depth analysis was performed on TC of B subtype, using ultra-deep sequencing (UDS) of env. Of 270 patients diagnosed with PHI during the study period, 229 were enrolled (median follow-up 168 (IQR 96-232) weeks). Median age: 39 (IQR 32-48) years; 94.8% males, 86.5% Italians, 83.4% MSM, 56.8% carrying HIV-1 subtype B. Of them, 92.6% started early treatment within a median of 4 (IQR 2-7) days after diagnosis; median time to sustained suppression was 20 (IQR 8-32) weeks. Twenty TC (median size 3, range 2-9 individuals), including 68 patients, were identified. A diagnosis prior to 2015 was the unique factor associated with inclusion in a TC. Added value of UDS was the identification of shared quasispecies components in transmission pairs within TC.
Keywords: HIV primary infection; phylogenetic analysis; spread and epidemiology; ultra-deep sequencing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Grant R.M., Lama J.R., Anderson P.L., Mcmahan V., Liu A.Y., Vargas L., Goicochea P., Casapía M., Guanira-Carranza J.V., Ramirez-Cardich M.E., et al. Preexposure Chemoprophylaxis for HIV Prevention in Men Who Have Sex with Men. N. Engl. J. Med. 2010;363:2587–2599. doi: 10.1056/NEJMoa1011205. - DOI - PMC - PubMed
-
- Gibson K.M., Jair K., Castel A.D., Bendall M.L., Wilbourn B., Jordan J.A., Crandall K.A., Pérez-Losada M., Subramanian T., Binkley J., et al. A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing. Sci. Rep. 2020;10:1989. doi: 10.1038/s41598-020-58410-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

