Coupling of a Novel TIMP3 Peptide to Carboxypeptidase G2 for Pro-Drug Activation at the Tumour Site
- PMID: 33504102
- PMCID: PMC7865317
- DOI: 10.3390/molecules26030625
Coupling of a Novel TIMP3 Peptide to Carboxypeptidase G2 for Pro-Drug Activation at the Tumour Site
Abstract
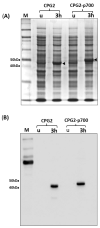
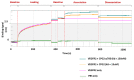
Broad-spectrum cytotoxic drugs have been used in cancer therapy for decades. However, their lack of specificity to cancer cells often results in serious side-effects, limiting efficacy. For this reason, antibodies have been used to attempt to specifically target cytotoxic drugs to tumours. One such approach is antibody-directed enzyme prodrug therapy (ADEPT) which uses a tumour-directed monoclonal antibody, coupled to an enzyme, to convert a systemically administered non-toxic prodrug into a toxic one only at the tumour site. Among the main drawbacks of ADEPT is the immunogenicity of the antibody-enzyme complex, which is exacerbated by slow clearance due to size, hence limiting repeated administration. Additionally, the mono-specificity of the antibody could potentially result in drug resistance with repeated administration. We have identified a novel short peptide sequence, p700, derived from a human tissue inhibitor of metalloproteinases-3 (TIMP-3), which binds to and inhibits a number of tyrosine kinase growth factor receptors (VEGFRs1-3, FGFRs 1-4 and PDGFRα) which are known to be upregulated in many tumours and tumour vasculature. In this report, we fused p700 to His-tagged, codon-optimised, carboxypeptidase G2 (CPG2). CPG2 is a bacterial enzyme used in ADEPT, which activates potent nitrogen-mustard pro-drugs by removal of an inhibitory glutamic acid residue. Recombinant CPG2-p700 was highly expressed in Escherichia coli and successfully purified by nickel affinity chromatography. Biolayer interferometry showed that CPG2-p700 had a 100-fold increase in binding affinity for VEGFR2 compared with CPG2 alone and retained its catalytic activity, as determined by methotrexate cleavage. In the presence of CPG2-p700, the ZD2676P pro-drug showed significant cytotoxicity for 4T1 cells compared with prodrug alone or CPG2 alone. p700 is, therefore, a potentially useful alternative to monoclonal antibodies for enzyme pro-drug therapy and could equally be used for effective delivery of other cytotoxic drugs to tumour tissue.
Keywords: TIMP3; VEGFR2; angiogenesis; carboxypeptidase G2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

