New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids
- PMID: 33504663
- PMCID: PMC7885322
- DOI: 10.1128/mSphere.01136-20
New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids
Abstract
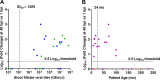
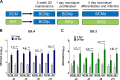
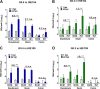
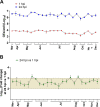
Human noroviruses (HuNoVs) are the leading cause of epidemic and sporadic acute gastroenteritis worldwide. We previously demonstrated human intestinal stem cell-derived enteroids (HIEs) support cultivation of several HuNoV strains. However, HIEs did not support virus replication from every HuNoV-positive stool sample, which led us to test and optimize new medium conditions, identify characteristics of stool samples that allow replication, and evaluate consistency of replication over time. Optimization of our HIE-HuNoV culture system has shown the following: (i) a new HIE culture medium made with conditioned medium from a single cell line and commercial media promotes robust replication of HuNoV strains that replicated poorly in HIEs grown in our original culture medium made with conditioned media from 3 separate cell lines; (ii) GI.1, 11 GII genotypes (GII.1, GII.2, GII.3, GII.4, GII.6, GII.7, GII.8, GII.12, GII.13, GII.14, and GII.17), and six GII.4 variants can be cultivated in HIEs; (iii) successful replication is more likely with virus in stools with higher virus titers; (iv) GII.4_Sydney_2012 virus replication was reproducible over 3 years; and (v) HuNoV infection is restricted to the small intestine, based on replication of two viral strains in duodenal and ileal HIEs, but not colonoids, from two susceptible donors. These results improve the HIE culture system for HuNoV replication. Use of HIEs by several laboratories worldwide to study the molecular mechanisms that regulate HuNoV replication confirms the usefulness of this culture system, and our optimized methods for virus replication will advance the development of effective therapies and methods for virus control.IMPORTANCE Human noroviruses (HuNoVs) are highly contagious and cause acute and sporadic diarrheal illness in all age groups. In addition, chronic infections occur in immunocompromised cancer and transplant patients. These viruses are antigenically and genetically diverse, and there are strain-specific differences in binding to cellular attachment factors. In addition, new discoveries are being made on strain-specific differences in virus entry and replication and the epithelial cell response to infection in human intestinal enteroids. Human intestinal enteroids are a biologically relevant model to study HuNoVs; however, not all strains can be cultivated at this time. A complete understanding of HuNoV biology thus requires cultivation conditions that will allow the replication of multiple strains. We report optimization of HuNoV cultivation in human intestinal enteroid cultures to increase the numbers of cultivatable strains and the magnitude of replication, which is critical for testing antivirals, neutralizing antibodies, and methods of virus inactivation.
Keywords: human intestinal enteroids; human norovirus; virus replication.
Copyright © 2021 Ettayebi et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

