IDH1 mutations induce organelle defects via dysregulated phospholipids
- PMID: 33504762
- PMCID: PMC7840755
- DOI: 10.1038/s41467-020-20752-6
IDH1 mutations induce organelle defects via dysregulated phospholipids
Abstract
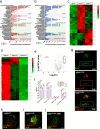
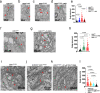
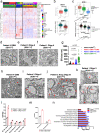
Infiltrating gliomas are devastating and incurable tumors. Amongst all gliomas, those harboring a mutation in isocitrate dehydrogenase 1 mutation (IDH1mut) acquire a different tumor biology and clinical manifestation from those that are IDH1WT. Understanding the unique metabolic profile reprogrammed by IDH1 mutation has the potential to identify new molecular targets for glioma therapy. Herein, we uncover increased monounsaturated fatty acids (MUFA) and their phospholipids in endoplasmic reticulum (ER), generated by IDH1 mutation, that are responsible for Golgi and ER dilation. We demonstrate a direct link between the IDH1 mutation and this organelle morphology via D-2HG-induced stearyl-CoA desaturase (SCD) overexpression, the rate-limiting enzyme in MUFA biosynthesis. Inhibition of IDH1 mutation or SCD silencing restores ER and Golgi morphology, while D-2HG and oleic acid induces morphological defects in these organelles. Moreover, addition of oleic acid, which tilts the balance towards elevated levels of MUFA, produces IDH1mut-specific cellular apoptosis. Collectively, these results suggest that IDH1mut-induced SCD overexpression can rearrange the distribution of lipids in the organelles of glioma cells, providing new insight into the link between lipid metabolism and organelle morphology in these cells, with potential and unique therapeutic implications.
Conflict of interest statement
The authors declare the following competing interests: P.P. is the owner of ACIS, LLC, a company developing BCAbox. A.K. is an employee of ACIS, LLC. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

