Salicylic diamines selectively eliminate residual undifferentiated cells from pluripotent stem cell-derived cardiomyocyte preparations
- PMID: 33504837
- PMCID: PMC7841182
- DOI: 10.1038/s41598-021-81351-z
Salicylic diamines selectively eliminate residual undifferentiated cells from pluripotent stem cell-derived cardiomyocyte preparations
Abstract
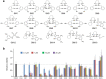
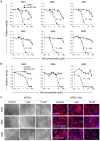
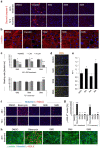
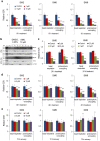
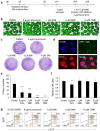
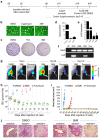
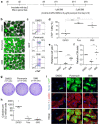
Clinical translation of pluripotent stem cell (PSC) derivatives is hindered by the tumorigenic risk from residual undifferentiated cells. Here, we identified salicylic diamines as potent agents exhibiting toxicity to murine and human PSCs but not to cardiomyocytes (CMs) derived from them. Half maximal inhibitory concentrations (IC50) of small molecules SM2 and SM6 were, respectively, 9- and 18-fold higher for human than murine PSCs, while the IC50 of SM8 was comparable for both PSC groups. Treatment of murine embryoid bodies in suspension differentiation cultures with the most effective small molecule SM6 significantly reduced PSC and non-PSC contamination and enriched CM populations that would otherwise be eliminated in genetic selection approaches. All tested salicylic diamines exerted their toxicity by inhibiting the oxygen consumption rate (OCR) in PSCs. No or only minimal and reversible effects on OCR, sarcomeric integrity, DNA stability, apoptosis rate, ROS levels or beating frequency were observed in PSC-CMs, although effects on human PSC-CMs seemed to be more deleterious at higher SM-concentrations. Teratoma formation from SM6-treated murine PSC-CMs was abolished or delayed compared to untreated cells. We conclude that salicylic diamines represent promising compounds for PSC removal and enrichment of CMs without the need for other selection strategies.
Conflict of interest statement
The use of the compounds described in this study for elimination of PSCs from their differentiated derivatives is covered by the Patent Application No. 18796954.8-1111. The authors A.B., T.Š., K.B. and J.H. are listed as inventors on this application. These authors declare no additional competing interests. The authors H.T., S.H., M.O., G.P., J.W.K., W.H. and K.P. declare no financial or non-financial conflicts of interest. Results described in this manuscript were used by K.B. in partial fulfillment of the requirements for the PhD degree at the University of Cologne.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

