Elucidating the tunability of binding behavior for the MERS-CoV macro domain with NAD metabolites
- PMID: 33504944
- PMCID: PMC7840908
- DOI: 10.1038/s42003-020-01633-6
Elucidating the tunability of binding behavior for the MERS-CoV macro domain with NAD metabolites
Abstract
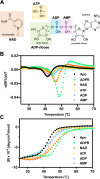
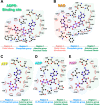
The macro domain is an ADP-ribose (ADPR) binding module, which is considered to act as a sensor to recognize nicotinamide adenine dinucleotide (NAD) metabolites, including poly ADPR (PAR) and other small molecules. The recognition of macro domains with various ligands is important for a variety of biological functions involved in NAD metabolism, including DNA repair, chromatin remodeling, maintenance of genomic stability, and response to viral infection. Nevertheless, how the macro domain binds to moieties with such structural obstacles using a simple cleft remains a puzzle. We systematically investigated the Middle East respiratory syndrome-coronavirus (MERS-CoV) macro domain for its ligand selectivity and binding properties by structural and biophysical approaches. Of interest, NAD, which is considered not to interact with macro domains, was co-crystallized with the MERS-CoV macro domain. Further studies at physiological temperature revealed that NAD has similar binding ability with ADPR because of the accommodation of the thermal-tunable binding pocket. This study provides the biochemical and structural bases of the detailed ligand-binding mode of the MERS-CoV macro domain. In addition, our observation of enhanced binding affinity of the MERS-CoV macro domain to NAD at physiological temperature highlights the need for further study to reveal the biological functions.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- MOST 109-2628-B-002-037/Ministry of Science and Technology, Taiwan (Ministry of Science and Technology of Taiwan)/International
- MOST 108-2113-M-002-011/Ministry of Science and Technology, Taiwan (Ministry of Science and Technology of Taiwan)/International
- MOST 109-2113-M-002-003/Ministry of Science and Technology, Taiwan (Ministry of Science and Technology of Taiwan)/International
- NTU-CC-L893501/National Taiwan University (NTU)/International
- NTU-109L7734/National Taiwan University (NTU)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

