Ebola virus antibody decay-stimulation in a high proportion of survivors
- PMID: 33505020
- PMCID: PMC7839293
- DOI: 10.1038/s41586-020-03146-y
Ebola virus antibody decay-stimulation in a high proportion of survivors
Abstract
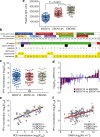
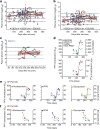
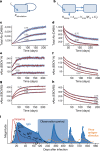
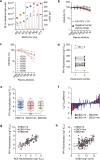
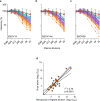
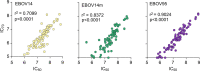
Neutralizing antibody function provides a foundation for the efficacy of vaccines and therapies1-3. Here, using a robust in vitro Ebola virus (EBOV) pseudo-particle infection assay and a well-defined set of solid-phase assays, we describe a wide spectrum of antibody responses in a cohort of healthy survivors of the Sierra Leone EBOV outbreak of 2013-2016. Pseudo-particle virus-neutralizing antibodies correlated with total anti-EBOV reactivity and neutralizing antibodies against live EBOV. Variant EBOV glycoproteins (1995 and 2014 strains) were similarly neutralized. During longitudinal follow-up, antibody responses fluctuated in a 'decay-stimulation-decay' pattern that suggests de novo restimulation by EBOV antigens after recovery. A pharmacodynamic model of antibody reactivity identified a decay half-life of 77-100 days and a doubling time of 46-86 days in a high proportion of survivors. The highest antibody reactivity was observed around 200 days after an individual had recovered. The model suggests that EBOV antibody reactivity declines over 0.5-2 years after recovery. In a high proportion of healthy survivors, antibody responses undergo rapid restimulation. Vigilant follow-up of survivors and possible elective de novo antigenic stimulation by vaccine immunization should be considered in order to prevent EBOV viral recrudescence in recovering individuals and thereby to mitigate the potential risk of reseeding an outbreak.
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Antibodies periodically wax and wane in survivors of Ebola.Nature. 2021 Feb;590(7846):397-398. doi: 10.1038/d41586-020-03044-3. Nature. 2021. PMID: 33504998 No abstract available.
References
-
- Kasangye, K. A. et al. in Advances in Ebola Control (ed. Okware, S. I.) (IntechOpen, 2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

