Whole-genome doubling confers unique genetic vulnerabilities on tumour cells
- PMID: 33505027
- PMCID: PMC7889737
- DOI: 10.1038/s41586-020-03133-3
Whole-genome doubling confers unique genetic vulnerabilities on tumour cells
Erratum in
-
Publisher Correction: Whole-genome doubling confers unique genetic vulnerabilities on tumour cells.Nature. 2021 May;593(7860):E15. doi: 10.1038/s41586-021-03591-3. Nature. 2021. PMID: 33976417 No abstract available.
Abstract
Whole-genome doubling (WGD) is common in human cancers, occurring early in tumorigenesis and generating genetically unstable tetraploid cells that fuel tumour development1,2. Cells that undergo WGD (WGD+ cells) must adapt to accommodate their abnormal tetraploid state; however, the nature of these adaptations, and whether they confer vulnerabilities that can be exploited therapeutically, is unclear. Here, using sequencing data from roughly 10,000 primary human cancer samples and essentiality data from approximately 600 cancer cell lines, we show that WGD gives rise to common genetic traits that are accompanied by unique vulnerabilities. We reveal that WGD+ cells are more dependent than WGD- cells on signalling from the spindle-assembly checkpoint, DNA-replication factors and proteasome function. We also identify KIF18A, which encodes a mitotic kinesin protein, as being specifically required for the viability of WGD+ cells. Although KIF18A is largely dispensable for accurate chromosome segregation during mitosis in WGD- cells, its loss induces notable mitotic errors in WGD+ cells, ultimately impairing cell viability. Collectively, our results suggest new strategies for specifically targeting WGD+ cancer cells while sparing the normal, non-transformed WGD- cells that comprise human tissue.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures

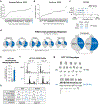



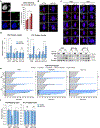
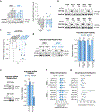





References
-
- Dewhurst SM et al. Tolerance of Whole-Genome Doubling Propagates Chromosomal Instability and Accelerates Cancer Genome Evolution. Cancer Discovery 4, 175, doi:10.1158/2159-8290.CD-13-0285 (2014). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

