Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition
- PMID: 33505028
- PMCID: PMC8262644
- DOI: 10.1038/s41586-020-03114-6
Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition
Abstract
Selective targeting of aneuploid cells is an attractive strategy for cancer treatment1. However, it is unclear whether aneuploidy generates any clinically relevant vulnerabilities in cancer cells. Here we mapped the aneuploidy landscapes of about 1,000 human cancer cell lines, and analysed genetic and chemical perturbation screens2-9 to identify cellular vulnerabilities associated with aneuploidy. We found that aneuploid cancer cells show increased sensitivity to genetic perturbation of core components of the spindle assembly checkpoint (SAC), which ensures the proper segregation of chromosomes during mitosis10. Unexpectedly, we also found that aneuploid cancer cells were less sensitive than diploid cells to short-term exposure to multiple SAC inhibitors. Indeed, aneuploid cancer cells became increasingly sensitive to inhibition of SAC over time. Aneuploid cells exhibited aberrant spindle geometry and dynamics, and kept dividing when the SAC was inhibited, resulting in the accumulation of mitotic defects, and in unstable and less-fit karyotypes. Therefore, although aneuploid cancer cells could overcome inhibition of SAC more readily than diploid cells, their long-term proliferation was jeopardized. We identified a specific mitotic kinesin, KIF18A, whose activity was perturbed in aneuploid cancer cells. Aneuploid cancer cells were particularly vulnerable to depletion of KIF18A, and KIF18A overexpression restored their response to SAC inhibition. Our results identify a therapeutically relevant, synthetic lethal interaction between aneuploidy and the SAC.
Conflict of interest statement
Competing Interests
T.R.G. is a consultant to GlaxoSmithKline and is a founder of Sherlock Biosciences. R.B. own shares in Ampressa and receives grant funding from Novartis. A.J.B. receives funding from Merck, Bayer and Novartis, and is an advisor to Earli and Helix Nano and a co-founder of Signet Therapeutics. The other authors declare no competing interests.
Figures



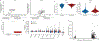
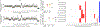









References
-
- Aguirre AJ et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. (2016) doi:10.1158/2159-8290.CD-16-0154. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

