Non-synonymous Substitutions in HIV-1 GAG Are Frequent in Epitopes Outside the Functionally Conserved Regions and Associated With Subtype Differences
- PMID: 33505382
- PMCID: PMC7829476
- DOI: 10.3389/fmicb.2020.615721
Non-synonymous Substitutions in HIV-1 GAG Are Frequent in Epitopes Outside the Functionally Conserved Regions and Associated With Subtype Differences
Abstract
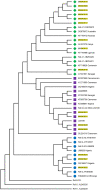
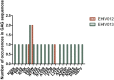
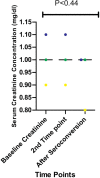
In 2019, 38 million people lived with HIV-1 infection resulting in 690,000 deaths. Over 50% of this infection and its associated deaths occurred in Sub-Saharan Africa. The West African region is a known hotspot of the HIV-1 epidemic. There is a need to develop an HIV-1 vaccine if the HIV epidemic would be effectively controlled. Few protective cytotoxic T Lymphocytes (CTL) epitopes within the HIV-1 GAG (HIV_gagconsv) have been previously identified to be functionally conserved among the HIV-1 M group. These epitopes are currently the focus of universal HIV-1 T cell-based vaccine studies. However, these epitopes' phenotypic and genetic properties have not been observed in natural settings for HIV-1 strains circulating in the West African region. This information is critical as the usefulness of universal HIV-1 vaccines in the West African region depends on these epitopes' occurrence in strains circulating in the area. This study describes non-synonymous substitutions within and without HIV_gagconsv genes isolated from 10 infected Nigerians at the early stages of HIV-1 infection. Furthermore, we analyzed these substitutions longitudinally in five infected individuals from the early stages of infection till after seroconversion. We identified three non-synonymous substitutions within HIV_gagconsv genes isolated from early HIV infected individuals. Fourteen and nineteen mutations outside the HIV_gagconsv were observed before and after seroconversion, respectively, while we found four mutations within the HIV_gagconsv. These substitutions include previously mapped CTL epitope immune escape mutants. CTL immune pressure likely leaves different footprints on HIV-1 GAG epitopes within and outside the HIV_gagconsv. This information is crucial for universal HIV-1 vaccine designs for use in the West African region.
Keywords: HIV cure; HIV-1 GAG; HIV-1 subtypes; MHR; non-synonymous substitutions.
Copyright © 2021 Olusola, Olaleye and Odaibo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa.J Immunol. 2004 Oct 1;173(7):4607-17. doi: 10.4049/jimmunol.173.7.4607. J Immunol. 2004. PMID: 15383595
-
Next-generation sequencing analyses of the emergence and maintenance of mutations in CTL epitopes in HIV controllers with differential viremia control.Retrovirology. 2018 Sep 10;15(1):62. doi: 10.1186/s12977-018-0444-z. Retrovirology. 2018. PMID: 30201008 Free PMC article.
-
Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef.J Infect Dis. 2005 Sep 1;192(5):749-59. doi: 10.1086/432547. Epub 2005 Jul 27. J Infect Dis. 2005. PMID: 16088824
-
Dendritic cells focus CTL responses toward highly conserved and topologically important HIV-1 epitopes.EBioMedicine. 2021 Jan;63:103175. doi: 10.1016/j.ebiom.2020.103175. Epub 2021 Jan 12. EBioMedicine. 2021. PMID: 33450518 Free PMC article.
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
Cited by
-
Accelerating Bayesian inference of dependency between mixed-type biological traits.PLoS Comput Biol. 2023 Aug 28;19(8):e1011419. doi: 10.1371/journal.pcbi.1011419. eCollection 2023 Aug. PLoS Comput Biol. 2023. PMID: 37639445 Free PMC article.
References
-
- Arcia D., Ochoa R., Hernández J. C., Cristiam M., Díaz F. J., Velilla P. A., et al. (2018). Potential immune escape mutations under inferred selection pressure in HIV-1 strains circulating in Medellin, Colombia. Infect. Genet. Evol. 69 267–278. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

