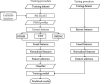
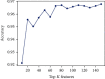
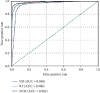
iT3SE-PX: Identification of Bacterial Type III Secreted Effectors Using PSSM Profiles and XGBoost Feature Selection
- PMID: 33505516
- PMCID: PMC7806399
- DOI: 10.1155/2021/6690299
iT3SE-PX: Identification of Bacterial Type III Secreted Effectors Using PSSM Profiles and XGBoost Feature Selection
Abstract
Identification of bacterial type III secreted effectors (T3SEs) has become a popular research topic in the field of bioinformatics due to its crucial role in understanding host-pathogen interaction and developing better therapeutic targets against the pathogens. However, the recognition of all effector proteins by using traditional experimental approaches is often time-consuming and laborious. Therefore, development of computational methods to accurately predict putative novel effectors is important in reducing the number of biological experiments for validation. In this study, we proposed a method, called iT3SE-PX, to identify T3SEs solely based on protein sequences. First, three kinds of features were extracted from the position-specific scoring matrix (PSSM) profiles to help train a machine learning (ML) model. Then, the extreme gradient boosting (XGBoost) algorithm was performed to rank these features based on their classification ability. Finally, the optimal features were selected as inputs to a support vector machine (SVM) classifier to predict T3SEs. Based on the two benchmark datasets, we conducted a 100-time randomized 5-fold cross validation (CV) and an independent test, respectively. The experimental results demonstrated that the proposed method achieved superior performance compared to most of the existing methods and could serve as a useful tool for identifying putative T3SEs, given only the sequence information.
Copyright © 2021 Chenchen Ding et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
Similar articles
-
PLM-T3SE: Accurate Prediction of Type III Secretion Effectors Using Protein Language Model Embeddings.J Cell Biochem. 2025 Jan;126(1):e30642. doi: 10.1002/jcb.30642. Epub 2024 Aug 20. J Cell Biochem. 2025. PMID: 39164870
-
ACNNT3: Attention-CNN Framework for Prediction of Sequence-Based Bacterial Type III Secreted Effectors.Comput Math Methods Med. 2020 Apr 3;2020:3974598. doi: 10.1155/2020/3974598. eCollection 2020. Comput Math Methods Med. 2020. PMID: 32328150 Free PMC article.
-
HMMPred: Accurate Prediction of DNA-Binding Proteins Based on HMM Profiles and XGBoost Feature Selection.Comput Math Methods Med. 2020 Mar 28;2020:1384749. doi: 10.1155/2020/1384749. eCollection 2020. Comput Math Methods Med. 2020. PMID: 32300371 Free PMC article.
-
Systematic analysis and prediction of type IV secreted effector proteins by machine learning approaches.Brief Bioinform. 2019 May 21;20(3):931-951. doi: 10.1093/bib/bbx164. Brief Bioinform. 2019. PMID: 29186295 Free PMC article.
-
Identify DNA-Binding Proteins Through the Extreme Gradient Boosting Algorithm.Front Genet. 2022 Jan 28;12:821996. doi: 10.3389/fgene.2021.821996. eCollection 2021. Front Genet. 2022. PMID: 35154264 Free PMC article. Review.
Cited by
-
Accurate Identification of Antioxidant Proteins Based on a Combination of Machine Learning Techniques and Hidden Markov Model Profiles.Comput Math Methods Med. 2021 Aug 7;2021:5770981. doi: 10.1155/2021/5770981. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34413898 Free PMC article.
-
PreAcrs: a machine learning framework for identifying anti-CRISPR proteins.BMC Bioinformatics. 2022 Oct 25;23(1):444. doi: 10.1186/s12859-022-04986-3. BMC Bioinformatics. 2022. PMID: 36284264 Free PMC article.
-
Natural language processing approach to model the secretion signal of type III effectors.Front Plant Sci. 2022 Oct 31;13:1024405. doi: 10.3389/fpls.2022.1024405. eCollection 2022. Front Plant Sci. 2022. PMID: 36388586 Free PMC article.
-
DeepT3 2.0: improving type III secreted effector predictions by an integrative deep learning framework.NAR Genom Bioinform. 2021 Oct 4;3(4):lqab086. doi: 10.1093/nargab/lqab086. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34617013 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources