Identified GNGT1 and NMU as Combined Diagnosis Biomarker of Non-Small-Cell Lung Cancer Utilizing Bioinformatics and Logistic Regression
- PMID: 33505535
- PMCID: PMC7806402
- DOI: 10.1155/2021/6696198
Identified GNGT1 and NMU as Combined Diagnosis Biomarker of Non-Small-Cell Lung Cancer Utilizing Bioinformatics and Logistic Regression
Abstract
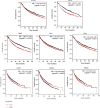
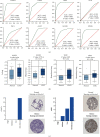
Non-small-cell lung cancer (NSCLC) is one of the most devastating diseases worldwide. The study is aimed at identifying reliable prognostic biomarkers and to improve understanding of cancer initiation and progression mechanisms. RNA-Seq data were downloaded from The Cancer Genome Atlas (TCGA) database. Subsequently, comprehensive bioinformatics analysis incorporating gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and the protein-protein interaction (PPI) network was conducted to identify differentially expressed genes (DEGs) closely associated with NSCLC. Eight hub genes were screened out using Molecular Complex Detection (MCODE) and cytoHubba. The prognostic and diagnostic values of the hub genes were further confirmed by survival analysis and receiver operating characteristic (ROC) curve analysis. Hub genes were validated by other datasets, such as the Oncomine, Human Protein Atlas, and cBioPortal databases. Ultimately, logistic regression analysis was conducted to evaluate the diagnostic potential of the two identified biomarkers. Screening removed 1,411 DEGs, including 1,362 upregulated and 49 downregulated genes. Pathway enrichment analysis of the DEGs examined the Ras signaling pathway, alcoholism, and other factors. Ultimately, eight prioritized genes (GNGT1, GNG4, NMU, GCG, TAC1, GAST, GCGR1, and NPSR1) were identified as hub genes. High hub gene expression was significantly associated with worse overall survival in patients with NSCLC. The ROC curves showed that these hub genes had diagnostic value. The mRNA expressions of GNGT1 and NMU were low in the Oncomine database. Their protein expressions and genetic alterations were also revealed. Finally, logistic regression analysis indicated that combining the two biomarkers substantially improved the ability to discriminate NSCLC. GNGT1 and NMU identified in the current study may empower further discovery of the molecular mechanisms underlying NSCLC's initiation and progression.
Copyright © 2021 Jia-Jia Zhang et al.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



Similar articles
-
Identification and validation of key genes with prognostic value in non-small-cell lung cancer via integrated bioinformatics analysis.Thorac Cancer. 2020 Apr;11(4):851-866. doi: 10.1111/1759-7714.13298. Epub 2020 Feb 14. Thorac Cancer. 2020. PMID: 32059076 Free PMC article.
-
Role of hemoglobin alpha and hemoglobin beta in non-small-cell lung cancer based on bioinformatics analysis.Mol Carcinog. 2022 Jun;61(6):587-602. doi: 10.1002/mc.23404. Epub 2022 Apr 8. Mol Carcinog. 2022. PMID: 35394695
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
MicroRNA-486: a dual-function biomarker for diagnosis and tumor immune microenvironment characterization in non-small cell lung cancer.BMC Med Genomics. 2025 May 19;18(1):92. doi: 10.1186/s12920-025-02158-9. BMC Med Genomics. 2025. PMID: 40390034 Free PMC article.
-
Logistic regression and other statistical tools in diagnostic biomarker studies.Clin Transl Oncol. 2024 Sep;26(9):2172-2180. doi: 10.1007/s12094-024-03413-8. Epub 2024 Mar 26. Clin Transl Oncol. 2024. PMID: 38530558 Free PMC article. Review.
Cited by
-
StackCirRNAPred: computational classification of long circRNA from other lncRNA based on stacking strategy.BMC Bioinformatics. 2022 Dec 27;23(1):563. doi: 10.1186/s12859-022-05118-7. BMC Bioinformatics. 2022. PMID: 36575368 Free PMC article.
-
High GNG4 expression is associated with poor prognosis in patients with lung adenocarcinoma.Thorac Cancer. 2022 Feb;13(3):369-379. doi: 10.1111/1759-7714.14265. Epub 2021 Dec 23. Thorac Cancer. 2022. PMID: 34951127 Free PMC article.
-
HPV, HBV, and HIV-1 Viral Integration Site Mapping: A Streamlined Workflow from NGS to Genomic Insights of Carcinogenesis.Viruses. 2024 Jun 18;16(6):975. doi: 10.3390/v16060975. Viruses. 2024. PMID: 38932267 Free PMC article.
-
Supervised Parametric Learning in the Identification of Composite Biomarker Signatures of Type 1 Diabetes in Integrated Parallel Multi-Omics Datasets.Biomedicines. 2024 Feb 22;12(3):492. doi: 10.3390/biomedicines12030492. Biomedicines. 2024. PMID: 38540105 Free PMC article.
-
A miR-212-3p/SLC6A1 Regulatory Sub-Network for the Prognosis of Hepatocellular Carcinoma.Cancer Manag Res. 2021 Jun 28;13:5063-5075. doi: 10.2147/CMAR.S308986. eCollection 2021. Cancer Manag Res. 2021. PMID: 34234551 Free PMC article.
References
-
- Ferlay J., Colombet M., Soerjomataram I., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. European Journal of Cancer. 2018;103:356–387. - PubMed
-
- Wang Z., Fu S., Zhao J., et al. Transbronchoscopic patient biopsy-derived xenografts as a preclinical model to explore chemorefractory-associated pathways and biomarkers for small-cell lung cancer. Cancer letters. 2019;440:180–188. - PubMed
-
- Han Y., Guo W., Ren T., et al. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer letters. 2019;440:116–125. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

