Down regulation of transcripts involved in selective metabolic pathways as an acclimation strategy in nitrogen use efficient genotypes of rice under low nitrogen
- PMID: 33505835
- PMCID: PMC7811496
- DOI: 10.1007/s13205-020-02631-5
Down regulation of transcripts involved in selective metabolic pathways as an acclimation strategy in nitrogen use efficient genotypes of rice under low nitrogen
Abstract
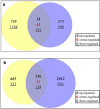
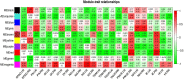
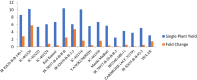
To understand the molecular mechanism of nitrogen use efficiency (NUE) in rice, two nitrogen (N) use efficient genotypes and two non-efficient genotypes were characterized using transcriptome analyses. The four genotypes were evaluated for 3 years under low and recommended N field conditions for 12 traits/parameters of yield, straw, nitrogen content along with NUE indices and 2 promising donors for rice NUE were identified. Using the transcriptome data generated from GS FLX 454 Roche and Illumina HiSeq 2000 of two efficient and two non-efficient genotypes grown under field conditions of low N and recommended N and their de novo assembly, differentially expressed transcripts and pathways during the panicle development were identified. Down regulation was observed in 30% of metabolic pathways in efficient genotypes and is being proposed as an acclimation strategy to low N. Ten sub metabolic pathways significantly enriched with additional transcripts either in the direction of the common expression or contra-regulated to the common expression were found to be critical for NUE in rice. Among the up-regulated transcripts in efficient genotypes, a hypothetical protein OsI_17904 with 2 alternative forms suggested the role of alternative splicing in NUE of rice and a potassium channel SKOR transcript (LOC_Os06g14030) has shown a positive correlation (0.62) with single plant yield under low N in a set of 16 rice genotypes. From the present study, we propose that the efficient genotypes appear to down regulate several not so critical metabolic pathways and divert the thus conserved energy to produce seed/yield under long-term N starvation.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-020-02631-5.
Keywords: Differential gene expression; Landraces; Metabolic pathways; Nitrogen use efficiency; SKOR transporter; Transcriptomics.
© King Abdulaziz City for Science and Technology 2021.
Conflict of interest statement
Conflict of interestAll the authors declare that they have no conflict of interest in the publication.
Figures
Similar articles
-
Rice Transcriptome Analysis Reveals Nitrogen Starvation Modulates Differential Alternative Splicing and Transcript Usage in Various Metabolism-Related Genes.Life (Basel). 2021 Mar 27;11(4):285. doi: 10.3390/life11040285. Life (Basel). 2021. PMID: 33801769 Free PMC article.
-
Enhanced expression of OsSPL14 gene and its association with yield components in rice (Oryza sativa) under low nitrogen conditions.Gene. 2016 Jan 15;576(1 Pt 3):441-50. doi: 10.1016/j.gene.2015.10.062. Epub 2015 Oct 28. Gene. 2016. PMID: 26519999
-
Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes.BMC Genomics. 2011 Oct 26;12:525. doi: 10.1186/1471-2164-12-525. BMC Genomics. 2011. PMID: 22029603 Free PMC article.
-
Promising physiological traits associated with nitrogen use efficiency in rice under reduced N application.Front Plant Sci. 2023 Nov 20;14:1268739. doi: 10.3389/fpls.2023.1268739. eCollection 2023. Front Plant Sci. 2023. PMID: 38053767 Free PMC article.
-
Molecular Regulatory Networks for Improving Nitrogen Use Efficiency in Rice.Int J Mol Sci. 2021 Aug 21;22(16):9040. doi: 10.3390/ijms22169040. Int J Mol Sci. 2021. PMID: 34445746 Free PMC article. Review.
Cited by
-
Genome-Wide Urea Response in Rice Genotypes Contrasting for Nitrogen Use Efficiency.Int J Mol Sci. 2023 Mar 23;24(7):6080. doi: 10.3390/ijms24076080. Int J Mol Sci. 2023. PMID: 37047052 Free PMC article.
-
Weighted gene co-expression network analysis of nitrogen (N)-responsive genes and the putative role of G-quadruplexes in N use efficiency (NUE) in rice.Front Plant Sci. 2023 Jun 7;14:1135675. doi: 10.3389/fpls.2023.1135675. eCollection 2023. Front Plant Sci. 2023. PMID: 37351205 Free PMC article.
-
Combined strategy employing MutMap and RNA-seq reveals genomic regions and genes associated with complete panicle exsertion in rice.Mol Breed. 2023 Aug 22;43(9):69. doi: 10.1007/s11032-023-01412-1. eCollection 2023 Sep. Mol Breed. 2023. PMID: 37622088 Free PMC article.
-
Identification and Characterization of Key Genes for Nitrogen Utilization from Saccharum spontaneum Sub-Genome in Modern Sugarcane Cultivar.Int J Mol Sci. 2024 Dec 30;26(1):226. doi: 10.3390/ijms26010226. Int J Mol Sci. 2024. PMID: 39796079 Free PMC article.
References
-
- Broadbent FE, De DSK, Laureles EV. Measurement of nitrogen utilization efficiency in rice genotypes. Agron J. 1987;791:786–791. doi: 10.2134/agronj1987.00021962007900050006x. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources