Apurinic/Apyrimidinic Endonuclease 1 and Tyrosyl-DNA Phosphodiesterase 1 Prevent Suicidal Covalent DNA-Protein Crosslink at Apurinic/Apyrimidinic Site
- PMID: 33505969
- PMCID: PMC7833210
- DOI: 10.3389/fcell.2020.617301
Apurinic/Apyrimidinic Endonuclease 1 and Tyrosyl-DNA Phosphodiesterase 1 Prevent Suicidal Covalent DNA-Protein Crosslink at Apurinic/Apyrimidinic Site
Abstract
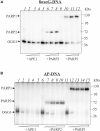
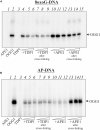
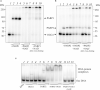
Bifunctional 8-oxoguanine-DNA glycosylase (OGG1), a crucial DNA-repair enzyme, removes from DNA 8-oxo-7,8-dihydroguanine (8-oxoG) with following cleavage of the arising apurinic/apyrimidinic (AP) site. The major enzyme in eukaryotic cells that catalyzes the cleavage of AP sites is AP endonuclease 1 (APE1). Alternatively, AP sites can be cleaved by tyrosyl-DNA phosphodiesterase 1 (TDP1) to initiate APE1-independent repair, thus expanding the ability of the base excision repair (BER) process. Poly(ADP-ribose) polymerase 1 (PARP1) is a regulatory protein of DNA repair. PARP2 is also activated in response to DNA damage and can be regarded as the BER participant. Here we analyze PARP1 and PARP2 interactions with DNA intermediates of the initial stages of the BER process (8-oxoG and AP-site containing DNA) and their interplay with the proteins recognizing and processing these DNA structures focusing on OGG1. OGG1 as well as PARP1 and PARP2 form covalent complex with AP site-containing DNA without borohydride reduction. AP site incision by APE1 or TDP1 removal of protein adducts but not proteins' PARylation prevent DNA-protein crosslinks.
Keywords: 8-oxoguanine-DNA glycosylase; AP endonuclease 1; DNA-protein crosslinks; apurinic/apyrimidinic site; poly(ADP-ribose) polymerases; tyrosyl-DNA phosphodiesterase 1.
Copyright © 2021 Lebedeva, Rechkunova, Endutkin and Lavrik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a shared affiliation and a past co-authorship with one of the authors OL at the time of review.
Figures

Similar articles
-
Apurinic/apyrimidinic endonuclease 1 has major impact in prevention of suicidal covalent DNA-protein crosslink with apurinic/apyrimidinic site in cellular extracts.IUBMB Life. 2024 Nov;76(11):987-996. doi: 10.1002/iub.2890. Epub 2024 Jul 4. IUBMB Life. 2024. PMID: 38963041
-
The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation.DNA Repair (Amst). 2018 Apr;64:10-25. doi: 10.1016/j.dnarep.2018.02.001. Epub 2018 Feb 11. DNA Repair (Amst). 2018. PMID: 29475157
-
Human apurinic/apyrimidinic endonuclease 1 is modified in vitro by poly(ADP-ribose) polymerase 1 under control of the structure of damaged DNA.Biochimie. 2020 Jan;168:144-155. doi: 10.1016/j.biochi.2019.10.011. Epub 2019 Oct 24. Biochimie. 2020. PMID: 31668992
-
[Tyrosyl-DNA Phosphodiesterase 1 Is a New Player in Repair of Apurinic/Apyrimidinic Sites].Bioorg Khim. 2015 Sep-Oct;41(5):531-8. doi: 10.1134/s106816201505012x. Bioorg Khim. 2015. PMID: 26762090 Review. Russian.
-
AP Endonuclease 1 as a Key Enzyme in Repair of Apurinic/Apyrimidinic Sites.Biochemistry (Mosc). 2016 Sep;81(9):951-67. doi: 10.1134/S0006297916090042. Biochemistry (Mosc). 2016. PMID: 27682167 Review.
Cited by
-
Targeting DNA Damage Repair and Immune Checkpoint Proteins for Optimizing the Treatment of Endometrial Cancer.Pharmaceutics. 2023 Aug 30;15(9):2241. doi: 10.3390/pharmaceutics15092241. Pharmaceutics. 2023. PMID: 37765210 Free PMC article. Review.
-
A Mini-Review of Flavone Isomers Apigenin and Genistein in Prostate Cancer Treatment.Front Pharmacol. 2022 Mar 11;13:851589. doi: 10.3389/fphar.2022.851589. eCollection 2022. Front Pharmacol. 2022. PMID: 35359832 Free PMC article. Review.
-
Human TDP1, APE1 and TREX1 repair 3'-DNA-peptide/protein cross-links arising from abasic sites in vitro.Nucleic Acids Res. 2022 Apr 22;50(7):3638-3657. doi: 10.1093/nar/gkac185. Nucleic Acids Res. 2022. PMID: 35349719 Free PMC article.
-
Targeting PARP proteins in acute leukemia: DNA damage response inhibition and therapeutic strategies.J Hematol Oncol. 2022 Jan 22;15(1):10. doi: 10.1186/s13045-022-01228-0. J Hematol Oncol. 2022. PMID: 35065680 Free PMC article. Review.
-
Identification of microRNA-mRNA regulatory network associated with oxidative DNA damage in human astrocytes.ASN Neuro. 2022 Jan-Dec;14:17590914221101704. doi: 10.1177/17590914221101704. ASN Neuro. 2022. PMID: 35570825 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

