Development of mPing-based activation tags for crop insertional mutagenesis
- PMID: 33506165
- PMCID: PMC7814626
- DOI: 10.1002/pld3.300
Development of mPing-based activation tags for crop insertional mutagenesis
Abstract
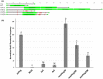
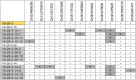
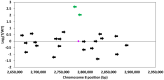
Modern plant breeding increasingly relies on genomic information to guide crop improvement. Although some genes are characterized, additional tools are needed to effectively identify and characterize genes associated with crop traits. To address this need, the mPing element from rice was modified to serve as an activation tag to induce expression of nearby genes. Embedding promoter sequences in mPing resulted in a decrease in overall transposition rate; however, this effect was negated by using a hyperactive version of mPing called mmPing20. Transgenic soybean events carrying mPing-based activation tags and the appropriate transposase expression cassettes showed evidence of transposition. Expression analysis of a line that contained a heritable insertion of the mmPing20F activation tag indicated that the activation tag induced overexpression of the nearby soybean genes. This represents a significant advance in gene discovery technology as activation tags have the potential to induce more phenotypes than the original mPing element, improving the overall effectiveness of the mutagenesis system.
Keywords: activation tagging; gene discovery; soybean; transposable element.
© 2021 The Authors. Plant Direct published by American Society of Plant Biologists, Society for Experimental Biology and John Wiley & Sons Ltd.
Figures



References
-
- Alonso, J. M. , Stepanova, A. N. , Leisse, T. J. , Kim, C. J. , Chen, H. M. , Shinn, P. , Stevenson, D. K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C. C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , … Ecker, J. R. (2003). Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. 10.1126/science.1086391 - DOI - PubMed
-
- Bechtold, N. , & Bouchez, D. (1995). In planta Agrobacterium‐mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration In Potrykus I. & Spangenberg G. (Eds.), Gene transfer to plants, Springer Lab Manual, (pp. 19–23). Berlin, Heidelberg: Springer; 10.1007/978-3-642-79247-2_3 - DOI
-
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

