Genotype-phenotype correlation identified a novel SARS-CoV-2 variant possibly linked to severe disease
- PMID: 33506644
- PMCID: PMC8013505
- DOI: 10.1111/tbed.14004
Genotype-phenotype correlation identified a novel SARS-CoV-2 variant possibly linked to severe disease
Abstract
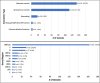
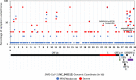
The geographic location and heterogeneous multi-ethnic population of Dubai (United Arab Emirates; UAE) provide a unique setting to explore the global molecular epidemiology of SARS-CoV-2 and relationship between different viral strains and disease severity. We systematically selected (i.e. every 100th individual in the central Dubai COVID-19 database) 256 patients by age, sex, disease severity and month to provide a representative sample of laboratory-confirmed COVID-19 patients (nasopharyngeal swab PCR positive) during the first wave of the UAE outbreak (January to June 2020). Sociodemographic and clinical data were extracted from medical records and full SARS-CoV-2 genome sequences extracted from nasopharyngeal swabs were analysed. Older age was significantly associated with COVID-19-associated hospital admission and mortality. Overweight/obese or diabetic patients were 3-4 times more likely to be admitted to hospital and intensive care unit (ICU). Sequencing data showed multiple independent viral introductions into the UAE from Europe, Iran and Asia (29 January-18 March), and these early strains seeded significant clustering consistent with almost exclusive community-based transmission between April and June 2020. Majority of sequenced strains (N = 60, 52%) were from the European cluster consistent with the higher infectivity rates associated with the D614G mutation carried by most strains in this cluster. A total of 986 mutations were identified in 115 genomes, 272 were unique (majority were missense, n = 134) and 20/272 mutations were novel. A missense (Q271R) and synonymous (R41R) mutation in the S and N proteins, respectively, were identified in 2/27 patients with severe COVID-19 but not in patients with mild or moderate disease (0/86; p = .05, Fisher's Exact Test). Both patients were women (51-64 years) with no significant underlying health conditions. The same two mutations were identified in a healthy 37-year-old Indian man who was hospitalized in India due to COVID-19. Our findings provide evidence for continued community-based transmission of the European strains in the Dubai population and highlight new mutations that might be associated with severe disease in otherwise healthy adults.
Keywords: COVID-19; Q271R; SARS-CoV-2; molecular phylogeny; mutation; whole genome sequencing.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
Authors have no conflicts of interest to disclose.
Figures


References
-
- Alm, E. , Broberg, E. K. , Connor, T. , Hodcroft, E. B. , Komissarov, A. B. , Maurer‐Stroh, S. , Melidou, A. , Neher, R. A. , O'Toole, Á. , Pereyaslov, D. & WHO European Region sequencing laboratories and GISAID EpiCoV group; WHO European Region sequencing laboratories and GISAID EpiCoV group* (2020). Geographical and temporal distribution of SARS‐CoV‐2 clades in the WHO European Region, January to June 2020. Euro Surveillance, 25, 2001410. 10.2807/1560-7917.ES.2020.25.32.2001410 - DOI - PMC - PubMed
-
- Butler, D. , Mozsary, C. , Meydan, C. , Dnako, D. , Foox, J. , Rosiene, J. , Shaiber, A. , Afshinnekoo, E. , MacKay, M. , Sedlazeck, F. J. , Ivanov, N. A. , Sierra, M. , Pohle, D. , Zietz, M. , Gisladottir, U. , Ramlall, V. , Westover, C. D. , Ryon, K. , Young, B. , … Mason, C. E. (2020). Shotgun transcriptome and isothermal profiling of SARS‐CoV‐2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv [Preprint], 2020.04.20.048066. 10.1101/2020.04.20.048066 - DOI - PMC - PubMed
-
- Harilal, D. , Ramaswamy, S. , Loney, T. , Al Suwaidi, H. , Khansaheb, H. , Alkhaja, A. , Varghese, R. , Deesi, Z. , Nowotny, N. , Alsheikh‐Ali, A. , & Tayoun, A. A. (2020). SARS‐CoV‐2 whole genome amplification and sequencing for effective population‐based surveillance and control of viral transmission. Clinical Chemistry, 66, 1450–1458. 10.1093/clinchem/hvaa187 - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

