The deubiquitylase USP9X controls ribosomal stalling
- PMID: 33507233
- PMCID: PMC7849821
- DOI: 10.1083/jcb.202004211
The deubiquitylase USP9X controls ribosomal stalling
Erratum in
-
Correction: The deubiquitylase USP9X controls ribosomal stalling.J Cell Biol. 2021 Mar 1;220(3):jcb.20200421102102021c. doi: 10.1083/jcb.20200421102102021c. J Cell Biol. 2021. PMID: 33600552 Free PMC article. No abstract available.
Abstract
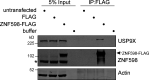
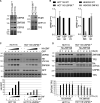
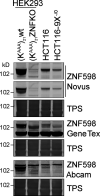
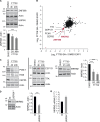
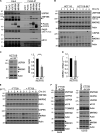
When a ribosome stalls during translation, it runs the risk of collision with a trailing ribosome. Such an encounter leads to the formation of a stable di-ribosome complex, which needs to be resolved by a dedicated machinery. The initial stalling and the subsequent resolution of di-ribosomal complexes requires activity of Makorin and ZNF598 ubiquitin E3 ligases, respectively, through ubiquitylation of the eS10 and uS10 subunits of the ribosome. We have developed a specific small-molecule inhibitor of the deubiquitylase USP9X. Proteomics analysis, following inhibitor treatment of HCT116 cells, confirms previous reports linking USP9X with centrosome-associated protein stability but also reveals a loss of Makorin 2 and ZNF598. We show that USP9X interacts with both these ubiquitin E3 ligases, regulating their abundance through the control of protein stability. In the absence of USP9X or following chemical inhibition of its catalytic activity, levels of Makorins and ZNF598 are diminished, and the ribosomal quality control pathway is impaired.
© 2021 Clancy et al.
Figures


References
-
- Altun, M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J., Kumar K.G., Konietzny R., Fischer R., Kogan E., et al. . 2011. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18:1401–1412. 10.1016/j.chembiol.2011.08.018 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

