Analysis of Rice Transcriptome Reveals the LncRNA/CircRNA Regulation in Tissue Development
- PMID: 33507446
- PMCID: PMC7843763
- DOI: 10.1186/s12284-021-00455-2
Analysis of Rice Transcriptome Reveals the LncRNA/CircRNA Regulation in Tissue Development
Abstract
Background: Long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) can play important roles in many biological processes. However, no study of the influence of epigenetics factors or the 3D structure of the genome in their regulation is available in plants.
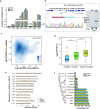
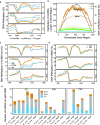
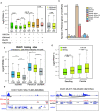
Results: In the current analysis, we identified a total of 15,122 lncRNAs and 7902 circRNAs in three tissues (root, leaf and panicle) in the rice varieties Minghui 63, Zhenshan 97 and their hybrid Shanyou 63. More than 73% of these lncRNAs and parental genes of circRNAs (P-circRNAs) are shared among Oryza sativa with high expression specificity. We found that, compared with protein-coding genes, the loci of these lncRNAs have higher methylation levels and the loci of circRNAs tend to locate in the middle of genes with high CG and CHG methylation. Meanwhile, the activated lncRNAs and P-circRNAs are mainly transcribed from demethylated regions containing CHH methylation. In addition, ~ 53% lncRNAs and ~ 15% P-circRNAs are associated with transposable elements (TEs), especially miniature inverted-repeat transposable elements and RC/Helitron. We didn't find correlation between the expression of lncRNAs and histone modifications; however, we found that the binding strength and interaction of RNAPII significantly affects lncRNA expression. Interestingly, P-circRNAs tend to combine active histone modifications. Finally, we found that lncRNAs and circRNAs acting as competing-endogenous RNAs have the potential to regulate the expression of genes, such as osa-156 l-5p (related to yield) and osa-miR444a-3p (related to N/P metabolism) confirmed through dual-luciferase reporter assays, with important roles in the growth and development of rice, laying a foundation for future rice breeding analyses.
Conclusions: In conclusion, our study comprehensively analyzed the important regulatory roles of lncRNA/circRNA in the tissue development of Indica rice from multiple perspectives.
Keywords: 3D genome; Circular RNAs (circRNAs); Competing endogenous RNAs (ceRNAs); DNA methylation; Epigenetic; Long non-coding RNAs (lncRNAs).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

