Aberrant Methylation of Tumour Suppressor Gene ADAM12 in Chronic Lympocytic Leukemia Patients: Application of Methylation Specific-PCR Technique
- PMID: 33507683
- PMCID: PMC8184192
- DOI: 10.31557/APJCP.2021.22.1.85
Aberrant Methylation of Tumour Suppressor Gene ADAM12 in Chronic Lympocytic Leukemia Patients: Application of Methylation Specific-PCR Technique
Abstract
Objective: Chronic Lymphocytic Leukemia (CLL) is a common leukemia among Caucasians but rare in Asians population. We postulated that aberrant methylation either hypermethylation or partial methylation might be one of the silencing mechanisms that inactivates the tumour suppressor genes in CLL. This study aimed to compare the methylation status of tumour suppressor gene, ADAM12, among CLL patients and normal individuals. We also evaluated the association between methylation of ADAM12 and clinical and demographic characteristics of the participants.
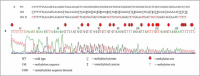
Methods: A total of 25 CLL patients and 25 normal individuals were recruited in this study. The methylation status of ADAM12 was determined using Methylation-Specific PCR (MSP); whereas, DNA sequencing method was applied for validation of the MSP results.
Results: Among CLL patients, 12 (48%) were partially methylated and 13 (52%) were unmethylated. Meanwhile, 5 (20%) and 20 (80.6%) of healthy individuals were partially methylated and unmethylated, respectively. There was a statistically significant association between the status of methylation at ADAM12 and the presence of CLL (p=0.037).
Conclusion: The aberrant methylation of ADAM12 found in this study using MSP assay may provide new exposure to CLL that may improve the gaps involved in genetic epigenetic study in CLL.
Keywords: ADAM12; Chronic Lympocytic Leukemia; methylation; tumour suppressor gene.
Figures


References
-
- Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Röcken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol. 2005;26:17–24. - PubMed
-
- Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. - PubMed
-
- Deligezer U, Erten N, Akisik EE, Dalay N. Methylation of the INK4A/ARF locus in blood mononuclear cells. Ann Hematol. 2006;85:102–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

