Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification
- PMID: 33508126
- PMCID: PMC8673443
- DOI: 10.1093/neuonc/noab012
Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification
Abstract
Background: Genome-wide DNA methylation profiling has recently been developed into a tool that allows tumor classification in central nervous system tumors. Extracellular vesicles (EVs) are released by tumor cells and contain high molecular weight DNA, rendering EVs a potential biomarker source to identify tumor subgroups, stratify patients and monitor therapy by liquid biopsy. We investigated whether the DNA in glioblastoma cell-derived EVs reflects genome-wide tumor methylation and mutational profiles and allows noninvasive tumor subtype classification.
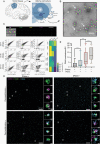
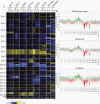
Methods: DNA was isolated from EVs secreted by glioblastoma cells as well as from matching cultured cells and tumors. EV-DNA was localized and quantified by direct stochastic optical reconstruction microscopy. Methylation and copy number profiling was performed using 850k arrays. Mutations were identified by targeted gene panel sequencing. Proteins were differentially quantified by mass spectrometric proteomics.
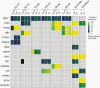
Results: Genome-wide methylation profiling of glioblastoma-derived EVs correctly identified the methylation class of the parental cells and original tumors, including the MGMT promoter methylation status. Tumor-specific mutations and copy number variations (CNV) were detected in EV-DNA with high accuracy. Different EV isolation techniques did not affect the methylation profiling and CNV results. DNA was present inside EVs and on the EV surface. Proteome analysis did not allow specific tumor identification or classification but identified tumor-associated proteins that could potentially be useful for enriching tumor-derived circulating EVs from biofluids.
Conclusions: This study provides proof of principle that EV-DNA reflects the genome-wide methylation, CNV, and mutational status of glioblastoma cells and enables their molecular classification.
Keywords: exosome; glioma; methylome; mutation; proteomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Osti D, Del Bene M, Rappa G, et al. . Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25(1):266–276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

