Cell competition constitutes a barrier for interspecies chimerism
- PMID: 33508854
- PMCID: PMC11163815
- DOI: 10.1038/s41586-021-03273-0
Cell competition constitutes a barrier for interspecies chimerism
Abstract
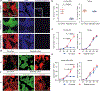
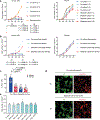
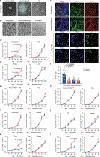
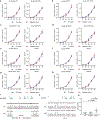
Cell competition involves a conserved fitness-sensing process during which fitter cells eliminate neighbouring less-fit but viable cells1. Cell competition has been proposed as a surveillance mechanism to ensure normal development and tissue homeostasis, and has also been suggested to act as a barrier to interspecies chimerism2. However, cell competition has not been studied in an interspecies context during early development owing to the lack of an in vitro model. Here we developed an interspecies pluripotent stem cell (PSC) co-culture strategy and uncovered a previously unknown mode of cell competition between species. Interspecies competition between PSCs occurred in primed but not naive pluripotent cells, and between evolutionarily distant species. By comparative transcriptome analysis, we found that genes related to the NF-κB signalling pathway, among others, were upregulated in less-fit 'loser' human cells. Genetic inactivation of a core component (P65, also known as RELA) and an upstream regulator (MYD88) of the NF-κB complex in human cells could overcome the competition between human and mouse PSCs, thereby improving the survival and chimerism of human cells in early mouse embryos. These insights into cell competition pave the way for the study of evolutionarily conserved mechanisms that underlie competitive cell interactions during early mammalian development. Suppression of interspecies PSC competition may facilitate the generation of human tissues in animals.
Conflict of interest statement
Figures









References
-
- Wu J. & Izpisua Belmonte JC Dynamic pluripotent stem cell states and their applications. Cell Stem Cell 17, 509–525 (2015). - PubMed
-
- Hackett JA & Surani MA Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15, 416–430 (2014). - PubMed
-
- Kobayashi T. et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

