Applying high throughput and comprehensive immunoinformatics approaches to design a trivalent subunit vaccine for induction of immune response against emerging human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2
- PMID: 33509045
- PMCID: PMC7852294
- DOI: 10.1080/07391102.2021.1876774
Applying high throughput and comprehensive immunoinformatics approaches to design a trivalent subunit vaccine for induction of immune response against emerging human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2
Abstract
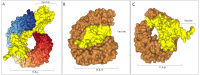
Coronaviruses (CoVs) cause diseases such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19). Therefore, this study was conducted to combat major CoVs via a trivalent subunit vaccine, which was engineered by implementing sequences of spike (S) protein, nucleocapsid (N), envelope (E), membrane (M) protein, non-structural protein (nsp) 3, and nsp8 antigens. The CTL, HTL, MHC I, and IFN-γ epitopes were predicted via CTLPRED, IEDB, and IFN epitope servers, respectively. Also, to stimulate strong helper T lymphocytes (HTLs) responses, Pan HLA DR-binding epitope (PADRE) was used. Also, for boosting the immune response, β-defensin 2 was added to the construct as an adjuvant. Furthermore, TAT was applied to the vaccine to facilitate the intracellular delivery. Finally, TAT, adjuvant, PADRE, and selected epitopes were appropriately assembled. Based on the predicted epitopes, a trivalent multi-epitope vaccine with a molecular weight of 74.8 kDa was constructed. Further analyses predicted the molecule to be a strong antigen, and a non-allergenic and soluble protein. Secondary and tertiary structures were predicted. Additionally, analyses validated the stability of the proposed vaccine. Molecular docking and molecular dynamics simulation (MDS) showed binding affinity and stability of the vaccine-TLR3 complex was favorable. The predicted epitopes demonstrated a strong potential to stimulate T and B-cell mediated immune responses. Furthermore, codon optimization and in silico cloning guaranteed increased expression. In summary, investigations demonstrated that this next-generation approach might provide a new horizon for the development of a highly immunogenic vaccine against SARS-CoV, MERS-CoV, and SARS-CoV-2.Communicated by Ramaswamy H. Sarma.
Keywords: Immunoinformatics; MERS‐CoV; SARS-CoV-2; SARS‐CoV; Subunit vaccine.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


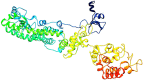




References
-
- Alexander, J., Fikes, J., Hoffman, S., Franke, E., Sacci, J., Appella, E., Chisari, F. V., Guidotti, L. G., Chesnut, R. W., Livingston, B., & Sette, A. (1998). The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunologic Research, 18(2), 79–92. 10.1007/BF02788751 - DOI - PubMed
-
- Azhar, E. I., Lanini, S., Ippolito, G., & Zumla, A. (2016). The Middle East respiratory syndrome coronavirus–a continuing risk to global health security. In Emerging and re-emerging viral infections. Advances in Experimental Medicine and Biology, 972, 49–60. 10.1007/5584_2016_133. - DOI - PMC - PubMed
-
- Biragyn, A., Coscia, M., Nagashima, K., Sanford, M., Young, H. A., & Olkhanud, P. (2008). Murine beta-defensin 2 promotes TLR-4/MyD88-mediated and NF-kappaB-dependent atypical death of APCs via activation of TNFR2. Journal of Leukocyte Biology, 83(4), 998–1008. 10.1189/jlb.1007700 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
